Figures & data
Figure 1. Alteration of glucose levels alters cell proliferation in a panel of human breast cancer cell lines. (A) The indicated human breast cancer cell lines were treated with various glucose concentrations (0, 5, 7, 10, 17 mmol/L) for 72 h then assayed for percent proliferation using MTS and normalized to 5 mmol/L glucose. (B–D) Indicated breast cancer cell lines used above were seeded in media containing 5 mmol/L (B), 10 mmol/L (C), or 17 mmol/L (D) glucose and treated with vehicle control, 5 mmol/L or 10 mmol/L metformin for 72 h. The percent cell proliferation was measured by MTS assay and normalized to control treatment. Experiments are representative of triplicate assays. Data are representative of averages of triplicate determinations ± SE.
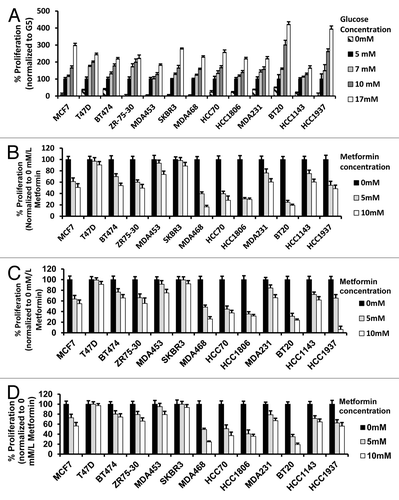
Table 1. EC50 for metformin at various glucose levels in human breast cancer cell lines
Figure 2. Reduction of glucose enhances metformin-mediated inhibition of colony formation and motility in TNBC, as compared with Luminal A or HER2 overexpressing cell lines. (A) Luminal A (T-47D), HER2 overexpressing (SK-BR-3), or TNBC (MDA-MB-468, MDA-MB-231) cell lines were treated with 5 mmol/L metformin in media containing various glucose levels (5, 10, 17 mmol/L) as described. (B) Enumeration of colony forming units for each cell line was performed see “Materials and Methods”. Bar graphs show averages of 3 independent experiments ± SE.
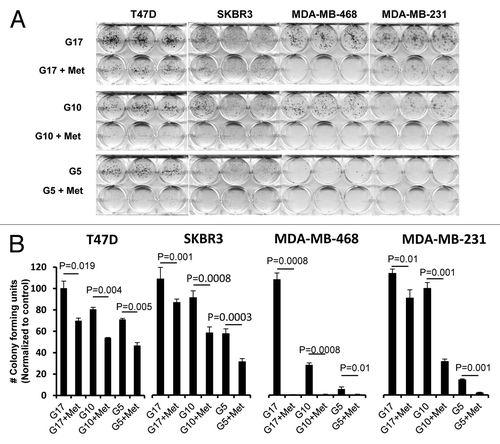
Figure 3. Lowering glucose levels enables metformin to effectively increase cellular apoptosis and decreases levels of cell cycle-regulated proteins. (A) Cell lines MCF7, SK-BR-3, MDA-MB-468, MDA-MB-231, and BT-549 were treated as described. Bar graphs show the percentage of cells in apoptosis (sub G1 fraction) are representative of 3 independent experiments ± SE. (B) Mod Fit analysis of flow cytometry studies performed in (A) shown with the addition of cells treated with 0 mmol/L glucose with or without metformin. Percentage of cells in apoptosis, G1, S, and G2/M are representative of 3 independent experiments. (C) MDA-MB-468 cells were treated as described and western blots performed for E2F1, Cyclins (A, B, D1, and E). Data shown are representative of 3 independent experiments.
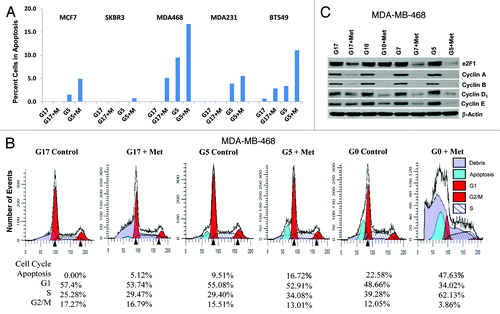
Figure 4. Metformin inhibits cellular motility and wound closure. (A) MDA-MB-231 cell were processed as described. Cover glass was stained by immol/Lunofluorescence (IF) for expression of moesin (green), F-actin (red), and DAPI (blue). Dotted line represents initial scratch at time zero. (B) Percent of wound closure of MDA-MB-231 (left) and MDA-MB-468 (right) was measured relative time 0; bar is representative of 40 μM. Columns are averages of (n = 6) determinants of 3 independent experiments ± SE. (C) MDA-MB-231 cells treated as described above were imaged at 100x on a Nikon microscope. Insets display colocalization of MSN/F-actin and filopodia extension of cells in 17 mmol/L glucose with or without 5 mmol/L metformin, bar is representative of 20 μM. Images are representative of 3 independent experiments.
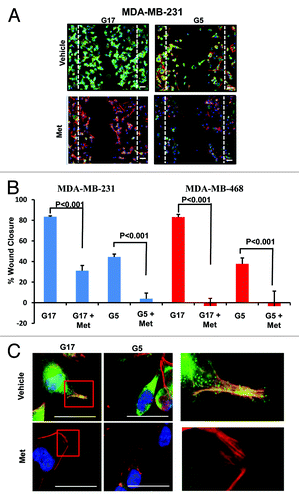
Figure 5. Physiological glucose enables metformin to effectively inhibit procarcinogenic signal transduction in SKBR3 and MDA-MB-468 cells. (A) SK-BR-3 (left) and MDA-MB-468 (right) cells treated with varying concentrations of glucose (5, 7, 10, or 17 mmol/L) in the absence or presence of 5 mmol/L metformin. Western blot membranes were probed for various signaling molecules as described in the “Materials and Methods”. Blots are representative of 3 independent experiments.
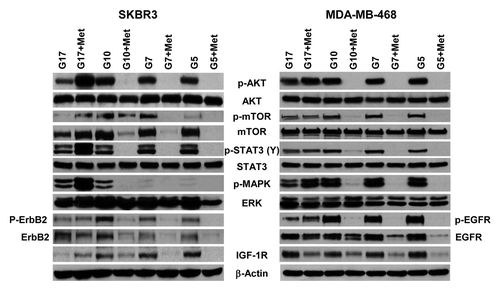
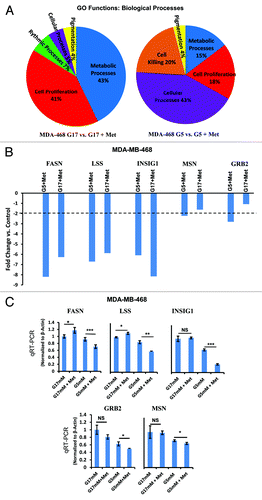
Figure 6. Microarray analysis of MDA-MB-468 cells cultured in media with normal (5 mmol/L) or high (17 mmol/L) glucose in the presence or absence of metformin. (A) MDA-MB-468 cells were treated as described and analyzed on the Affymetrix Human Gene 1.0 ST Array platform. Gene groups differentially expressed from gene ontology (GO) functions for biological processes as a result of above treatment are shown as a pie chart and representative of biological triplicates. (B) Selected genes differentially regulated by metformin in 5 or 17 mmol/L glucose as determined by Affymetrix gene array. Columns are representative of fold change relative to controls, and are averages of biological triplicate determinants ± SE. (C) Quantitative RT-PCR was performed on MDA-MB-468 treated as indicated above normalized to β-actin control. Columns are representative of triplicate determinants ± SE.