Figures & data
Figure 1. Temporal and spatial pattern of RBBP7 expression during oocyte maturation and preimplantation embryo development. (A) Immunocytochemical analysis of RBBP7 expression. All samples were processed for immunocytochemistry together, and all images were taken at the same laser power. The experiment was conducted 3 times, and at least 20 oocytes/embryos were analyzed for each sample. Shown are representative examples. The scale bar represents 50 μm. (B) Immunoblot analysis of RBBP7 expression. Forty oocytes/embryos were loaded per lane, and the experiment was conducted twice, and β-actin (ACTB) was used as a loading control. Shown is a representative example. (C) Corresponding histogram of relative immunoblot analysis and the data are expressed as mean ± SEM. GV, germinal vesicle; Met I, metaphase I; Met II, metaphase II; 1C, 1-cell embryo; 2C, 2-cell embryo; 8C, 8-cell embryo; BL, blastocyst.
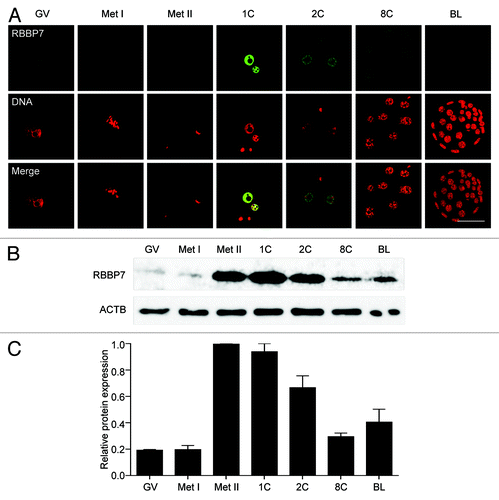
Figure 2. siRNA and morpholino mediated knockdown of RBBP7. (A) Immunoblot analysis of RBBP7 expression. Full-grown oocytes were injected with combination of siRNA and morpholino. After 18 h, 40 eggs were used for immunoblot analysis and β-actin (ACTB) was used as a loading control. The experiment was performed 6×. Shown is a representative example. Met II-u is 40, uninjected eggs; Met II-c is 40 control injected eggs. (B) Corresponding histogram of relative immunoblot analysis of (A). One-way ANOVA was used to analyze the data. The data are expressed as mean ± SEM. Values with an asterisk vary significantly, P < 0.05.
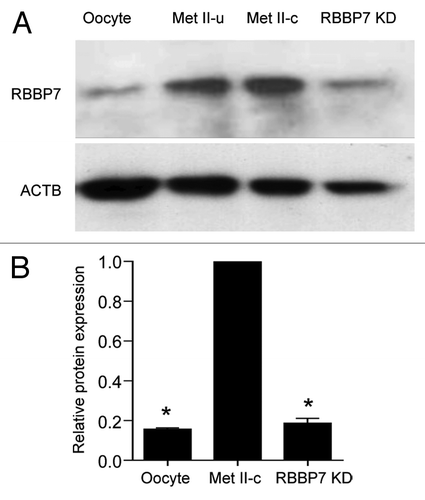
Figure 3. Effect of RBBP7 knockdown on histone acetylation status in MII oocytes. Immunocytochemical detection of H3K4ac (A), H4K8ac (B), H4K12ac (C), and H4K16ac (D) in RBBP7 KD Met II eggs. The experiments were performed 3×, and at least 20 oocytes were analyzed for each sample. The scale bars represent 10 μm. Shown are representative examples. The graphs on the right of the images are average quantifications of the pixel intensity of the indicated histone marks to the left. The data are expressed as mean ± SEM; Student t test was used to analyze the data. Values with an asterisk vary significantly, P < 0.05.
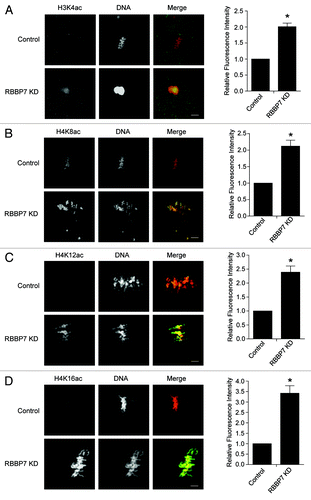
Figure 4. Effect of RBBP7 knockdown on meiotic progression. (A–C) Full-grown oocytes were injected with combination of siRNA and morpholino followed by in vitro maturation for 18 h and scored for first polar body extrusion (A) followed by fixation and staining with an anti-β-tubulin antibody (green) to detect spindles and TOPRO-3 to detect DNA (red) using confocal microscopy (B). The confocal images were analyzed for the presence of misaligned chromosomes and to detect abnormal spindle morphology. (C) MII eggs were treated with monastrol for 2 h prior to fixation and detection of kinetochores with Crest anti-serum and DNA with TOPRO-3. Eggs with greater or less than 40 kinetochores were considered aneuploid. The experiment was performed 3×, and at least 30 oocytes were examined for each sample. The scale bar represents 100 μm. The data are expressed as mean ± SEM; Student t test was used to analyze the data. Values with asterisks vary significantly, *P < 0.05, **P < 0.01, ****P < 0.0001.
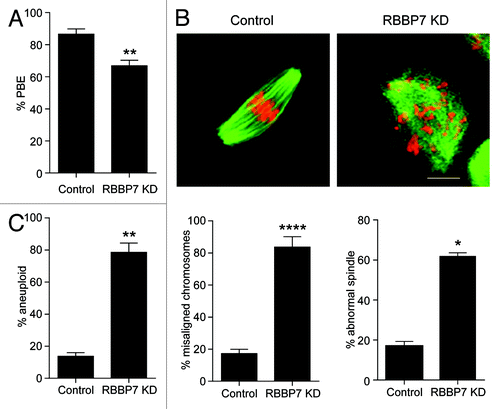
Figure 5. Time-lapse confocal observations of meiosis following RBBP7 knockdown. (A) Full-grown oocytes were injected with combination of siRNA and morpholino, and cRNAs encoding H2b-mCherry (red) and Aurka-Gfp (green) followed by in vitro maturation for 18 h. Bright field and fluorescence photographs were acquired every 20 min. Examples of observed abnormal cytokinesis are noted in Telo I. The scale bar represents 50 μm. (B) Quantification of abnormal cytokinesis defects in (A). GV, germinal vesicle intact oocyte; Pro-Met I, prophase I-metaphase I); Ana I, anaphase I; Telo I, telophase I; Met II, metaphase II. The data are expressed as mean ± SEM; Student t test was used to analyze the data. Values with asterisks vary significantly, *P < 0.05, ***P < 0.001.
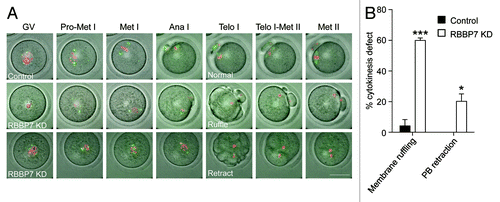
Figure 6. Effect of RBBP7 knockdown on spindle formation, chromosome alignment, and K-MT attachment. (A) Microtubules (green) and DNA (red) from the indicated treatment group were stained at Met I and examined for spindle formation and chromosome alignment by immunocytochemistry. The experiment was conducted 3×, and at least 20 oocytes were analyzed for each sample. The scale bar represents 10 μm. Shown are representative examples. (B) Corresponding quantification of (A). Student t test was used to analyze the data. (C) Representative confocal images from the cold-stable microtubule assay from the indicated treatment groups. Kinetochores were detected with Crest anti-sera (red), microtubules with an anti-β-tubulin antibody (green) and DNA with DAPI (blue). The insets highlight normal (control panel), abnormal (KD panel 1), or unattached (KD panel 2) kinetochores. The scale represents 10 μm. (D) Quantification of the types of attachments observed. At least 750 kinetochores were analyzed from 2 different experiments. The data are expressed as mean ± SEM; One-way ANOVA was used to analyze the data. Values with asterisks vary significantly, ***P < 0.001, ****P < 0.0001.
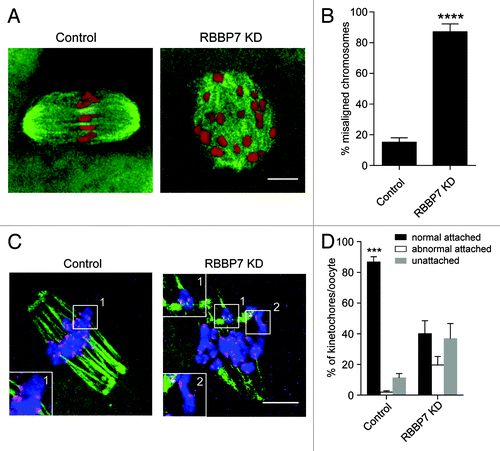
Figure 7. Effect of RBBP7 knockdown on the spindle assembly checkpoint. (A) Full-grown oocytes were injected with combination of siRNA and morpholino. Control-injected oocytes were incubated in maturation medium plus/minus 400 nM nocadozole (Noc). After 16 h of in vitro maturation, extrusion of the polar body was confirmed using light microscopy. The data are expressed as mean ± SEM; One-way ANOVA was used to analyze the data. (B) Representative examples of live imaging of oocytes from the indicated treatment groups co-injected with H2b-mCherry cRNA. The arrows point to lagging chromosomes. The scale bar represents 10 μm. (C) Quantification of (B). The data are expressed as mean ± SEM; Student t test was used to analyze the data. Values with asterisks vary significantly, *P < 0.05, ***P < 0.001.
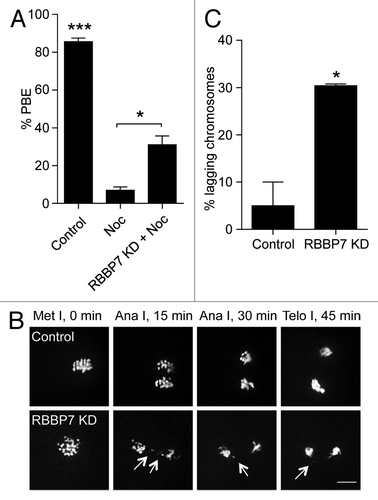
Figure 8. Effect of RBBP7 knockdown on the CPC during MI. Immunocytochemical detection of AURKC (A and B), Survivin (C), and pINCENP (D) (green in merge) in control and RBBP7 KD Met I oocytes. DNA was detected with DAPI (red in merge). In (B), 100 ng/μl AURKC-GFP (green in merge) was co-injected with the siRNA/morpholino cocktail. To the right of the image panels are quantifications of the intensity levels (A and D) or the percent of oocytes where normal chromosome localization was observed (B and C). The experiments were performed 2×, and at least 15 oocytes were analyzed for each sample. The scale bar represents 10 μm. Shown are representative examples. The data are expressed as mean ± SEM; Student t test was used to analyze the data. Values with asterisks vary significantly, * P < 0.05, **P < 0.01.
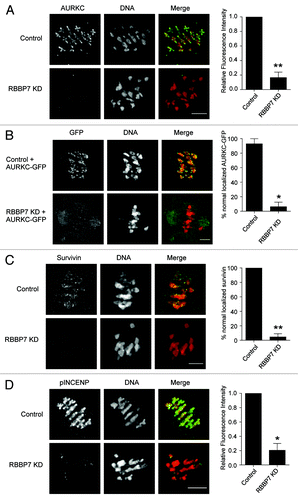
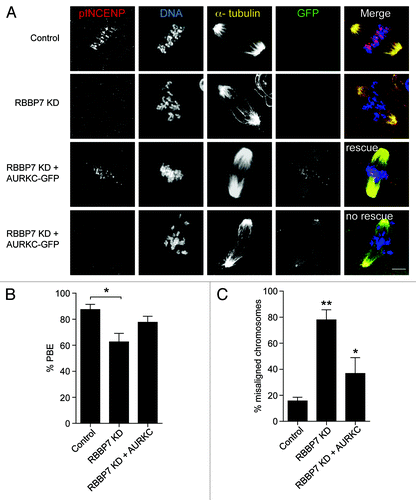
Figure 9. Overexpression of AURKC rescues the defects observed in RBBP7 knockdown oocytes. Full-grown oocytes were injected with the indicated materials and matured in vitro for 18 h, prior to fixation and detection of phosphorylated INCENP (pINCENP; red), the spindle (α-tubulin; yellow), and DNA (DAPI; blue). (A) Representative confocal images of each treatment group. In the “rescue” group, AURKC-GFP signal was found at kinetochores (green), whereas in the “no rescue” group, AURKC-GFP signal was at the spindle poles. Scale bar represents 10 μm. (B and C) Quantification of the percentage of oocytes in (A) that extruded a polar body (PBE), and had chromosome misalignment, respectively. The experiments were performed 3×, and at least 15 oocytes were analyzed for each sample. The data are expressed as mean ± SEM; One-way ANOVA was used to analyze the data. Values with asterisks vary significantly, * P < 0.05, **P < 0.01.