Figures & data
Figure 1. Defective enrichment of the PML (pro-myelocytic leukemia) protein on E2F-responsive cell cycle gene promoters in senescent Rb1∆L/∆L cells. Asynchronously growing wild-type and Rb1∆L/∆L MEFs were induced to senesce by retroviral-mediated expression of oncogenic HrasV12. Chromatin from proliferating and senescent cells was used for chromatin immunoprecipitation. Real-time PCR was used to amplify the immunoprecipitated DNA using primers specific to Cyclin E1 (left) and Mcm3 (right). The quantity of precipitated DNA is represented as percent of input chromatin. (A) ChIP of proliferating and senescent cells of the indicated genotypes using a α-H3K9me3 antibody or an IgG control. (B) ChIP of chromatin from proliferating and senescent wild-type and Rb1∆L/∆L cells using a α-PML antibody and an IgG control. (C) ChIP on senescent wild-type and Rb1∆L/∆L cells using a α-pRB antibody and an IgG control. All experiments were reproduced in at least 3 independent pairs of MEFs. Error bars indicate one standard deviation from the mean of at least 3 biological replicates. An asterisk indicates a statistically significant difference (t test, P < 0.05).
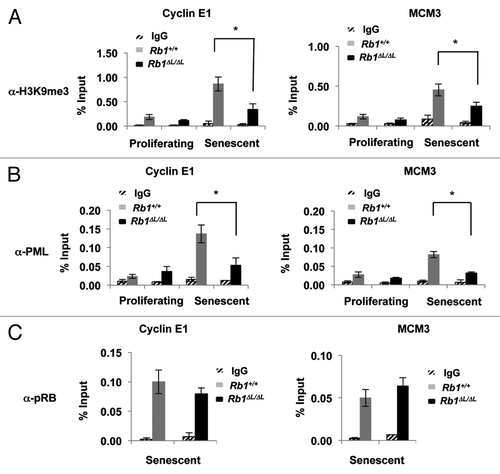
Figure 2. Oncogenic Ras induces DNA damage and accumulation of PML nuclear bodies in both wild-type and Rb1∆L/∆L cells. Asynchronously growing wild-type and Rb1∆L/∆L MEFs were induced to senesce by retroviral-mediated expression of oncogenic HrasV12. After 3 d of selection, cells were re-plated and cultured for the indicated amount of time. (A) Immunofluorescent (IF) staining of wild-type and Rb1∆L/∆L cells with γH2AX antibody (red) to detect double strand breaks at different times after induction of senescence. Nuclei were counter stained with DAPI (blue). (B) Quantification of DNA damage foci in (A). The percent of nuclei with >3 γH2AX foci were compared between genotypes. (C) Whole-cell extracts from asynchronously growing or oncogenic HrasV12 expressing senescent wild-type and Rb1∆L/∆L MEFs were probed with a γH2AX antibody. β-actin levels are used as loading control. (D) IF staining for PML nuclear bodies using a α-PML antibody (green) and DNA counterstaining with DAPI (blue). Inset images show detailed PML staining of individual nuclei. (E) Quantification of the number of PML bodies per nucleus in (D). The proportion of cells with fewer than 10, 10 to 25, or more than 25 PML bodies per nucleus are displayed in graphical format. All experiments were reproduced in at least 3 independent pairs of MEFs. Error bars represent one standard deviation from the mean of at least 3 biological replicates. Scale bars are 50 μM.
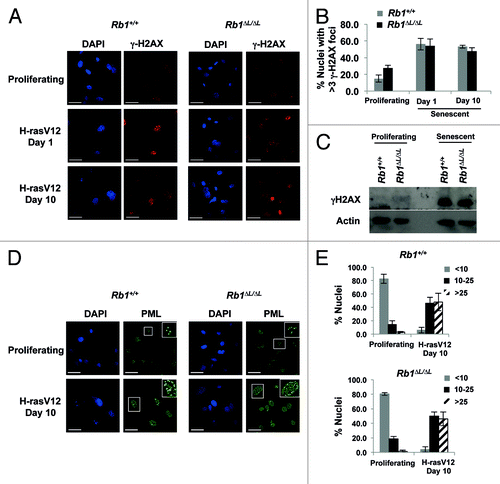
Figure 3. Defective senescent arrest in Rb1∆L/∆L MEFs expressing PML-IV. Asynchronously growing wild-type and Rb1∆L/∆L MEFs were transduced with retroviruses expressing pBABE-FLAG-PML-IV. After 3 d of drug selection, cells were plated in medium and cultured for the indicated amount of time. (A) Immunofluorescent (IF) staining was performed with a α-FLAG antibody (green) to detect PML-IV. Nuclei were counter stained with DAPI (blue). The inset image shows higher magnification of individual nuclei. (B) Cells of the indicated genotypes were pulsed with BrdU for 4 h, followed by fixation and staining with α-BrdU antibodies. The percentage of BrdU-positive nuclei at the indicated time points following FLAG-PML-IV expression are plotted. (C) PML-IV-expressing cells were stained for senescence-associated β-galactosidase (SA-β-gal) expression 8 d after the expression of PML-IV. (D) The number of SA-β-gal-positive cells in each genotype were quantified and plotted. (E) Quantification of E2F target gene mRNA from wild-type and Rb1∆L/∆L MEFs either from proliferating (left), or PML-IV-expressing senescent cells (right). Samples were normalized to expression of the ribosomal protein gene Rplp0. (F) Western blots to determine the expression of protein products of E2F target genes (p107 and cyclin E) following PML expression are shown. (G) DNA synthesis in response to ectopic E2F1 expression was measured by BrdU incorporation. Two days following Ad-E2F1 infection, cells were pulse labeled with BrdU for 16hrs and positive cells were identified by immunofluorescence microscopy and shown in the graph (bottom left). The fold increase in BrdU incorporation between control and E2F1 infected cells was calculated for both the genotypes and is shown in the graph on the bottom right. The mean fold increase was compared by a t test. The expression level of ectopic E2F1 in both the wild-type and Rb1∆L/∆L MEFs is shown by western blotting along with β-actin as a loading control. An asterisk on a graph indicates a statistically significant difference (t test, P < 0.05), an asterisk on a blot represents a non-specific band. All experiments were reproduced in at least three independent pairs of MEFs. Error bars represent one standard deviation from the mean of at least 3 biological replicates. Scale bars are 50 μM.
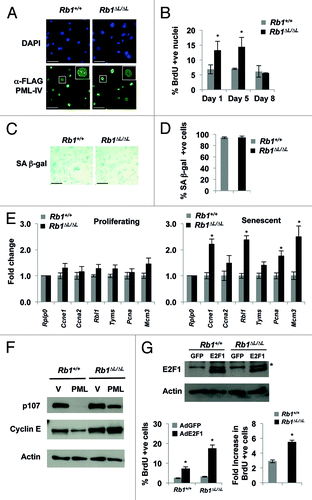
Figure 4. The pRB∆L mutation disrupts PML-pRB interactions during senescence. GST pull-down experiments were performed using nuclear extracts from proliferating or senescent MEFs induced to senesce by expression of oncogenic HrasV12. (A) GST pull-down using GST tagged pRB large pocket or pRB large pocket with ∆LXCXE mutations (∆L). GST alone is used as a negative control. Pull-down fractions were probed with antibodies specific to murine PML and E2F3. (B) GST pull-down as in (A) using GST tagged p107 large pocket. Pull-down fractions were probed with antibodies specific to either murine PML or E2F4. (C) Nuclear extracts from wild-type and Rb1∆L/∆L MEFs that were induced to senesce by expressing oncogenic HrasV12 were used for co-immunoprecipitation using a sheep α-RB antibody or a non-specific IgG control. The immunoprecipitated fractions were probed with a monoclonal antibody against PML (Millipore). The blots were then stripped and reprobed for E2F3. An asterisk indicates a non-specific band. (D) Nuclear extracts from proliferating and senescent cells were analyzed by SDS-PAGE and western blotting with a pan PML antibody that recognizes numerous isoforms (SC, shown at left). The same extracts were probed with a murine specific antibody to PML as above (MP, shown to the right). The arrows indicate differentially expressed bands. (E) GST pull-downs were performed as in (A), except the blot was probed with the pan PML antibody that recognizes many PML isoforms. Arrows indicate different PML species that are sensitive to ∆L mutations in pRB. Stars indicate cross reactivity with the GST-RB protein. (F) MEFs were transfected with expression constructs for each of the indicated PML isoforms. Following SDS-PAGE and western blotting, membranes were probed with the same pan PML antibody as in (E) to identify the migration pattern of different PML isoforms.
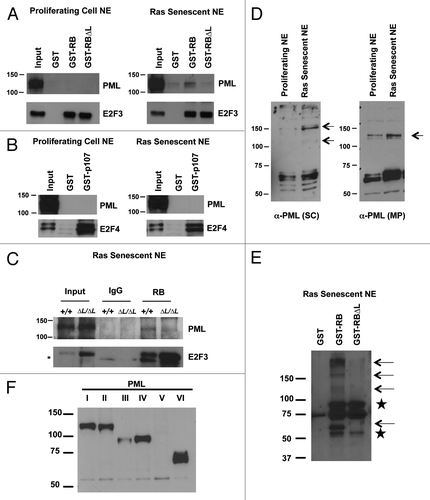
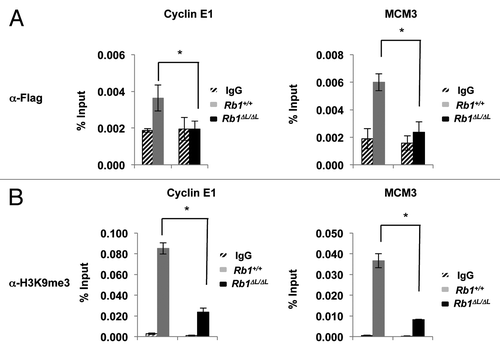
Figure 5. Defective enrichment of Flag-PML-IV and heterochromatin formation at E2F target gene promoters in Rb1∆L/∆L MEFs. Asynchronously growing wild-type and Rb1∆L/∆L MEFs were transduced with pBABE-FLAG-PML-IV retrovirus. After 3 d of drug selection, cells were re-plated and cultured for 8 more days before processing for chromatin immunoprecipitation. (A) ChIP on wild-type and Rb1∆L/∆L MEFs expressing PML-IV using a α-FLAG antibody or an IgG control. Real-time PCR was used to amplify the immunoprecipitated DNA using primers specific to the promoter regions of Ccne1 (left) and Mcm3 (right). (B) ChIP on wild-type and Rb1∆L/∆L MEFs expressing PML-IV using a α-H3K9me3 antibody or an IgG control. Real-time PCR was used to amplify the immunoprecipitated DNA using primers specific to the promoter regions of Ccne1 and Mcm3. All experiments were reproduced in at least 3 independent pairs of MEFs. Error bars represent one standard deviation from the mean of at least 3 biological replicates. An asterisk indicates a statistically significant difference (t test, P < 0.05).