Figures & data
Figure 1. Overview of CRC classification studies. (A and B) Graphic showing the clinical and biological characteristics and gene signatures of Colon Cancer Subtypes (CCS) (A) and CRCassigner subtypes (B). SSA, sessile serrated adenoma; TA, transit-amplifying; CR-TA, cetuximab-resistant TA; CS-TA, cetuximab-sensitive TA; DFS, disease-free survival.
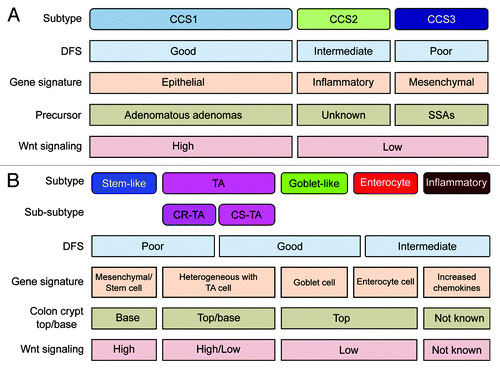
Figure 2. Relationships of colorectal cancer subtype classifications. (A and B) Heatmaps depict the AMC-AJCCII-90 data set classified according to the CRCassigner signature (A) and the Sadanandam et al. core data set classified based on the CCS classification (B). Columns represent patients; rows indicate CRCassigner genes (A) or CCS classifier genes (B). Colors represent relative gene expression levels; blue signifies low expression, and brown high expression. (C and D) Heatmaps indicate association of CRCassigner subtype samples (left) with CCS group samples (top) for the AMC-AJCCII-90 set (C) and the Sadanandam et al. core data set (D). Colors indicate significance of association; green signifies a low association, and red a high association. P values are determined using hypergeometric tests.
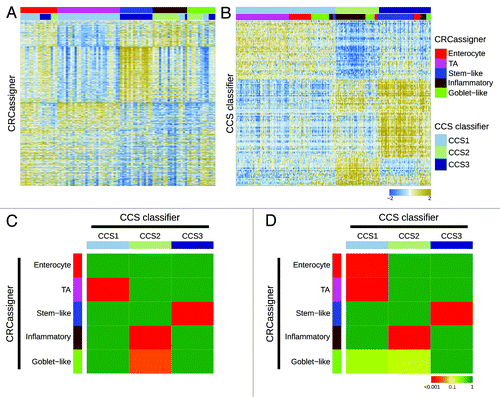
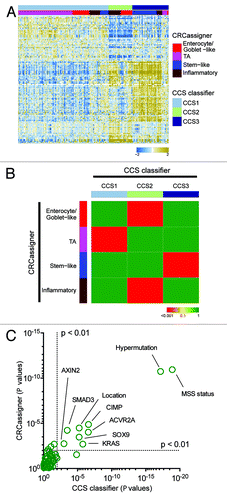
Figure 3. Validation in the TCGA data set. (A) Heatmap depicts the TCGA data set classified by both taxonomies. Columns represent patients; rows indicate genes from the CCS classifier. Colors represent relative gene expression levels; blue signifies low expression, and brown, high expression. (B) Heatmap indicates association of CRCassigner subtype samples (left) with CCS group samples (top) for the TCGA set. Colors indicate significance of association; green signifies a low association, and red a high association. P values are determined using hypergeometric tests. (C) Graph depicts the association of clinical and (epi)genetic characteristics with both classification methods for the TCGA data set. Features that are significantly associated with both classification schemes are indicated. P values are determined using Chi-square tests.