Figures & data
Figure 1.CENP-E or Nsl1 depletion induces aneuploidy and massive tissue overgrowth upon additional blockade of the apoptotic pathway. (A–H) Wing discs expressing MyrT, p35, and dsRNA forms of the indicated genes under the control of the ap-gal4 driver (labeled in red), and stained for DAPI (blue or white). Larvae were 6 d (A–D), 5 d (E), 11 d (F), 8 d (G) and 9-d-old (H). All pictures were taken at the same magnification. Bar in (A–H) represents 50 µm. (I) DNA content analysis by fluorescence-activated cell sorting (FACS) of dorsal (left panels) and ventral cells (right panels) of wing primordia shown in (B and C), respectively. Percentage of cells with DNA content higher than 4n is indicated. Genotypes: ap-gal4,UAS-myrT/+; UAS-p35/+ (A), ap-gal4,UAS-myrT/+; UAS-CENP-ERNAi/UAS-p35 (B and F),ap-gal4,UAS-myrT/UAS-Nsl1RNAi; UAS-p35/+ (C and G), ap-gal4,UAS-myrT/UAS–Nsl1RNAi(GD); UAS-p35/+ (D and H), ap-gal4,UAS-myrT/+; UAS-p35/ UAS-CENP-ERNAi(NIG) (E). d, days.
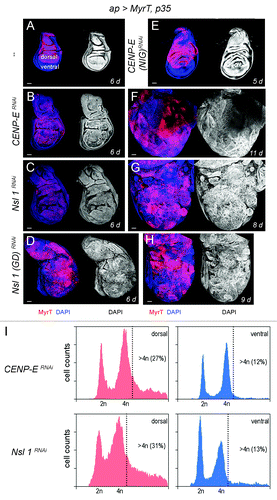
Figure 2.CENP-E- or Nsl1-depleted tissues overgrow and invade neighboring tissues upon additional blockade of the apoptotic pathway. (A–E) Wing primordia expressing the indicated transgenes in posterior cells under the control of the en-gal4 driver, and stained for DAPI (blue), Ci (red) and p35 (green). Larvae were 5 d (A–C), 6 d (D), and 12-d-old (E). All pictures were taken at the same magnification. Bar in (A–E) represents 50 µm. Ci labels the anterior (A) compartment. (F) Histogram plotting the P/A size ratio of wing primordia shown in (A–C). Genotypes: en-gal4,UAS-p35/+ (A); en-gal4,UAS-p35/+; UAS-CENP-ERNAi/+ (B and D); en-gal4,UAS-p35/UAS-Nsl1RNAi (C); en-gal4,UAS-p35/UAS–Nsl1RNAi(GD) (E). d, days.
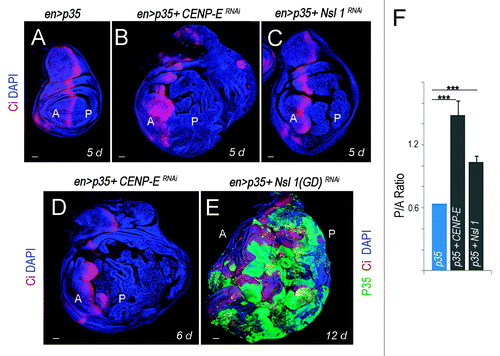
Figure 3. Depletion of CENP-E or Nsl1 drives tumor-like growth in allograft transplants. (A–C) Micrographs of adult flies carrying MyrT-labeled (red) implants of the indicated genotypes. Pictures were taken 12 d after implantation, and ratios indicating the reproducibility of the phenotype are shown. (A’–C”) Allograft transplants of the indicated genotypes stained to visualize MMP1 (a'–c', blue), Wg (A”–C”, blue), MyrT (red) expression, and DAPI (blue or white). Transplants were extracted 12 d after implantation. Bar in (a’–c”) represents 50 µm. Genotypes: ap-gal4,UAS-myrT/+; UAS-p35/+ (A–A”); ap-gal4,UAS-myrT/+; UAS-CENP-ERNAi/UAS-p35 (B–B”); ap-gal4,UAS-myrT/UAS-Nsl1RNAi; UAS-p35/+ (C–C”). d, days.
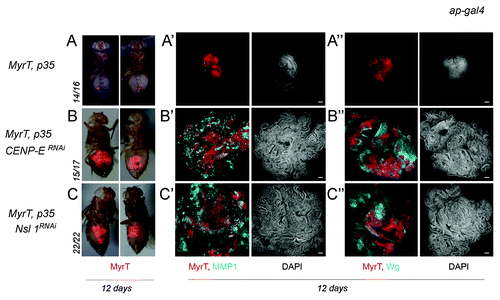
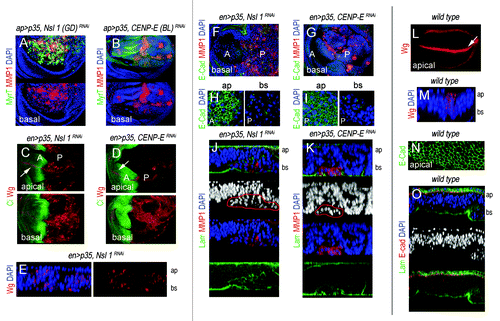
Figure 4. Cell delamination, Wingless, and MMP1 expression, and basement membrane degradation in CENP-E- or Nsl1-depleted tissues. (A and B) Wing primordia expressing the indicated transgenes under the control of the ap-gal4 driver, and stained for MyrT (green), MMP1 (red) and DAPI (blue). Basal sides of the epithelia are shown. (C and D) Wing primordia expressing the indicated transgenes under the control of the en-gal4 driver, and stained for Wg (red), and Ci (green). Ci labels the anterior (A) compartment. Ectopic expression of Wg was mainly observed in the basal side of the epithelium. White arrows indicate the endogenous expression of Wg along the future wing margin. (E) Cross-section of a wing pirmordium expressing the indicated transgenes under the control of the en-gal4 driver and stained for DAPI (blue) and Wg (red). ap, apical, and bs, basal. (F and G) Wing primordia expressing the indicated transgenes under the control of the en-gal4 driver, and stained for DAPI (blue), E-cad (green), and MMP1 (red). (H and I) Magnification of the apical (ap) and basal (bs) sections of the wing primordial are shown in (F and G). In (C, D, and F–I), anterior (A) and posterior (P) compartments are indicated. (J and K) Cross-sections of the posterior compartment of wing primordia expressing the indicated transgenes under the control of the en-gal4 driver and stained for DAPI (blue), laminin-γ (labels the BM, in green), and MMP1 (red). Expression of MMP1 is restricted to delaminated cells (marked in red). ap, apical; bs, basal. (L) Wild-type wing primordium stained for Wg (red). White arrow indicates the endogenous expression of Wg along the future wing margin. (M) Cross-section of a wild-type wing pirmordium stained for DAPI (blue), and Wg (red). ap, apical; bs, basal. (N) Magnification of the apical section of a wild-type wing pirmordium stained for E-cad (green) expression. (O) Cross-section of a wild-type wing pirmordium stained for DAPI (blue), laminin-γ (green), and E-cad (red). ap, apical; bs, basal. Genotypes: ap-gal4,UAS-myrT/UAS–Nsl1RNAi(GD); UAS-p35/+ (A), ap-gal4,UAS-myrT/+, UAS-p35/ UAS-CENP-ERNAi(BL) (B), en-gal4,UAS-p35/UAS-Nsl1RNAi (C, E, F, H, and J); en-gal4,UAS-p35/+; UAS-CENP-ERNAi/+ (D, G, I, and K), wild–type (L–O).