Figures & data
Figure 1. Western blot analysis of phosphorylated GR, LC3, SQSTM1, and GAPDH in L6 myotubes treated with Dex for indicated time points (A). LC3 and Hoechst 33342 immunofluorescence in L6 cells incubated with Dexamethasone for 6 h (B). Western blot analysis of LC3, SQSTM1, and GAPDH in L6 myotubes incubated with Baf A1 and/or Dex for 6 h (C). Western blot analysis of phophorylated MTOR, total MTOR, phosphorylated AMPK, total AMPK, LC3, SQSTM1, and GAPDH in L6 myotubes incubated with Dexamethasone for 0, 6, and 24 h (D). Western blot analysis of LC3, SQSTM1, BECN1, and GAPDH in LUC and BECN1 knockout L6 myotubes (E). Western blot analysis of GR, LC3, SQSTM1, and GAPDH in control and glucocorticoid receptor knockdown L6 myotubes (F). Data: mean ± SEM of at least 3 independent experiments. Statistically significant differences were calculated using ANOVA in combination with a Tukey test for group comparison. *P < 0.05 vs. control.
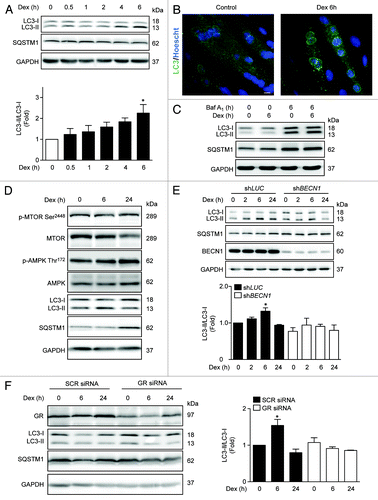
Figure 2. Western blot analysis of LC3, SQSTM1 and GAPDH in Dex, and/or Act D and CHX incubated L6 myotubes for indicated time points (A). Quantification of SQSTM1/GAPDH presented in (A) (B). Quantification of ATG5, SQSTM1, LC3, and BECN1 mRNA levels of control and Dexamethasone incubated L6 myotubules (C). Western blot analysis of LC3-II, SQSTM1, phosphorylated and total AMPK and GAPDH in compound C treated L6 myotubes incubated with Dex for indicated time points (D). Western blot analysis of LC3-II, SQSTM1, AMPKα1, and GAPDH in SCR and AMPKα1 knockdown L6 myotubes incubated with Dex for indicated time points (E). Data: mean ± SEM of at least 3 independent experiments. Statistically significant differences were calculated using ANOVA in combination with a Tukey test for group comparison. *P < 0.05 vs. control, #P < 0.05 vs. Dex.
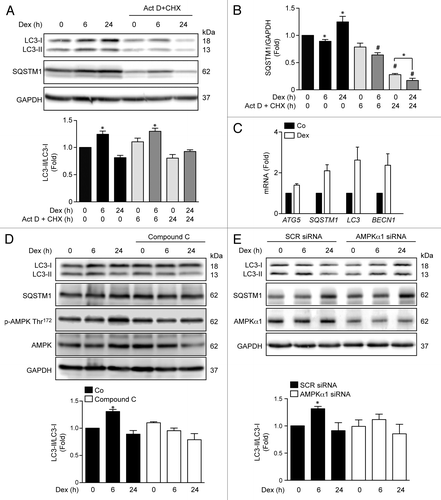
Figure 3. Mitochondria morphology shown by confocal microscopy in L6 myoblast incubated with Dex for 0, 6, and 24h and quantification of mitochondrial volume and number per cell (A). Western blot analysis of DNM1L, MFN2, mtHSP70, and GAPDH in Dex treated L6 myotubes for 0, 6, and 24 h (B). Histamine induced Ca2+ release in control and Dex-treated L6 myotubes (C). Histamine induced Ca2+ uptake by mitochondria in control and Dex incubated L6 myotubes (D). Data: mean ± SEM of at least 3 independent experiments. Statistically significant differences were calculated using ANOVA in combination with a Tukey test for group comparison. *P < 0.05 vs. control.
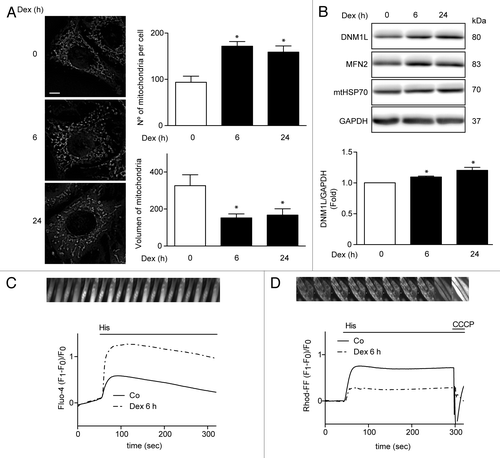
Figure 4. Mitochondria membrane potential (measured by TMRM fluorescence) (A), ROS induction (measured by Dihydrorhodamine-123 fluorescence) (B), oxygen consumption (C), and ATP levels (D) of L6 myotubes incubated with Dex for 0, 6, and 24 h. Data: mean ± SEM of at least 3 independent experiments. Statistically significant differences were calculated using ANOVA in combination with a Tukey test for group comparison. *P < 0.05 vs. control.
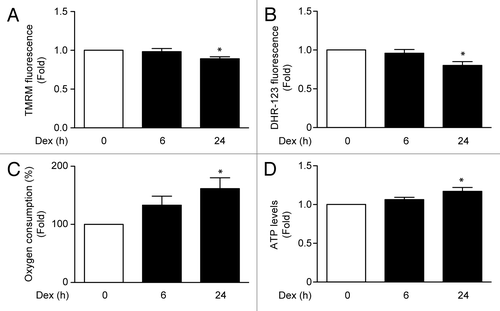
Figure 5. Mitochondria morphology shown by confocal microscopy in SCR and AMPKα1 knockdown L6 myotubes incubated with Dex for 0, 6, and 24 h (A). Quantification of mitochondrial volume and number per cell presented in (A). (B) Mitochondria morphology shown by confocal microscopy in LUC and BECN1 knockout L6 myotubes incubated with Dex for 0, 6, and 24 h (C). Quantification of mitochondrial volume and number per cell presented in (C). (D) Quantification of mitochondrial volume and number per cell in SCR and BECN1 knockdown L6 myotubes incubated with Dex for 0, 6, and 24 h (E). Western blot analysis of DNM1L, BECN1, LC3, and GAPDH in SCR and BECN1 knockdown L6 myotubes incubated with Dex for 0, 6, and 24 h (F). Data: mean ± SEM of at least 3 independent experiments. Statistically significant differences were calculated using ANOVA in combination with a Tukey test for group comparison. *P < 0.05 vs. time 0 (SCR siRNA). #P < 0.05 vs. SCR siRNA + Dex.
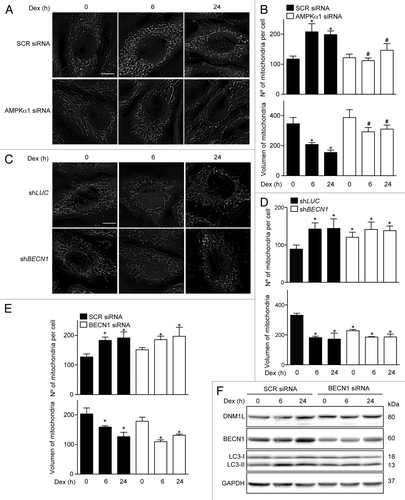
Figure 6. Mitochondria (MTG) and lysosome (LSTR) colocalization shown by confocal microscopy in control, Dex. and Dex plus Mdivi-1 (DNM1L inhibition) incubated L6 myoblast, quantification of mitochondria and lysosome colocalization expressed by Manders’ coefficient in L6 myoblast incubated with Dex and/or Mdivi-1 for 0, 6, and 24 h (A). Western blot analysis of PINK1, PARK2, and GAPDH in L6 myotubes incubated with Dex and/or Mdivi-1 for 0, 6, and 24 h (B). Oxygen consumption of Dex and/or Mdivi-1 incubated L6 myotubes (C). Western blot analysis of DNM1L, MFN2, mtHSP70, phophorylated AMPK, total AMPK, LC3, SQSTM1, and GAPDH in Dex and/or Mdivi-1 incubated L6 myotubes (D). Western blot analysis of LC3, SQSTM1 and GAPDH in Dex, Mdivi-1, and/or Baf A1 treated L6 myotubes (E). Data: mean ± SEM of at least 3 independent experiments. Statistically significant differences were calculated using ANOVA in combination with a Tukey test for group comparison. *P < 0.05 vs. control, #P < 0.05 vs. Dex.
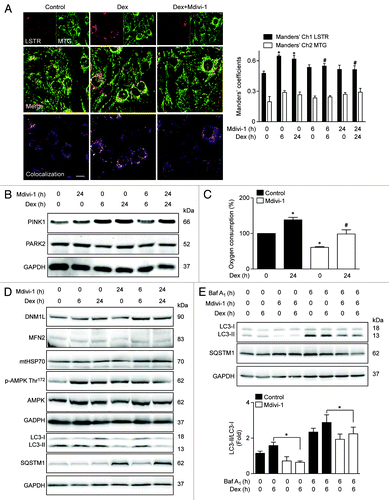
Figure 7. Quantification of ATG5, SQSTM1, LC3, and BECN1 mRNA levels of control, Dex, Mdivi-1, and Dex plus Mdivi-1 incubated L6 myotubes (A). Quantification of MURF1 (B) and ATROGIN-1 (C) mRNA levels in control and Mdivi-1 incubated L6 myotubes. Data: mean ± SEM of at least 3 independent experiments. Statistically significant differences were calculated using ANOVA in combination with a Tukey test for group comparison. *P < 0.05 vs. control, #P < 0.05 vs. Dex.
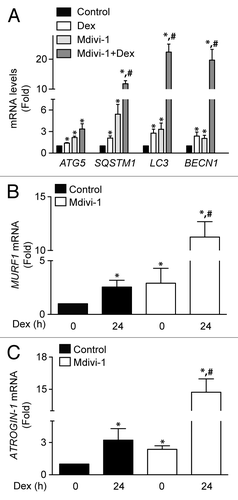
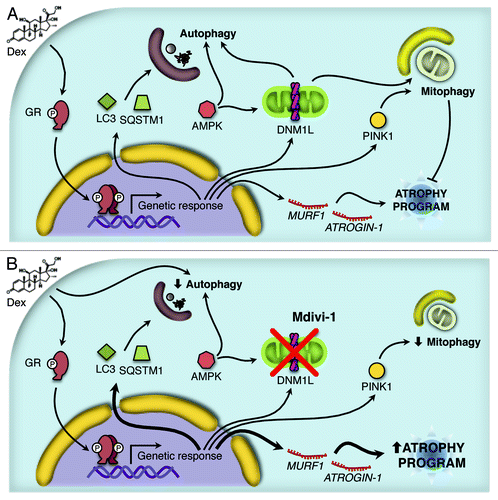
Figure 8. Working model proposed for the Dex action. Dex induces activation of the autophagy process through the protein AMPK and the expression of several autophagy-related genes and the atrophy program. In addition, Dex triggers mitochondrial fragmentation and consequent mitophagy (A). The inhibition of DNM1L by Mdivi-1 disrupts the mitochondrial fragmentation, autophagy and mitophagy induced by Dex. Furthermore, there is an increment in Dex-induced autophagy-related genes and the atrophy program as consequent of the autophagy/mitophagy reduction (B).