Figures & data
Chart 1. Chemical structures of the allosteric inhibitors 1–7 and those resulting from the hit expansion of compound 4.
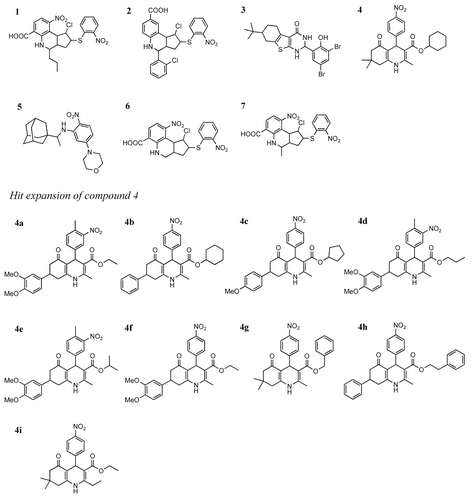
Table 1. Predicted free energies of binding, VS rankings, chemical classification, occupation of the ANS1 and ANS2 pockets, selected physico–chemical properties, and biological properties of the 7 allosteric inhibitors discovered in the primary virtual screening (compounds 1-7) and of the nine analogs of compound 4 selected from hit expansion (compounds 4a-4i)
Figure 1. Concentration-dependent displacement of ANS from CDK2. Panel (A) reports the displacement activity of compounds 1–7, while (B) shows the activity of six (4b–4g) of the nine compounds derived from the hit expansion of compound 4. Compounds 4a, 4h, and 4i did not show appreciable displacement activity.
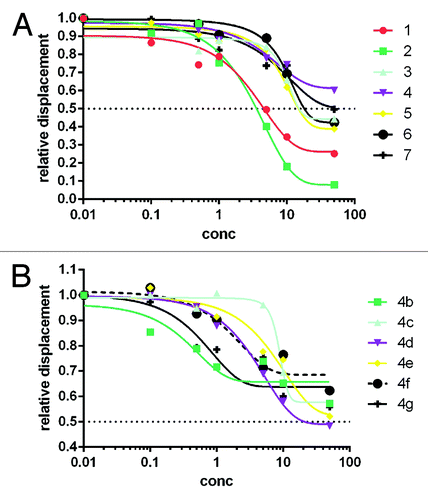
Figure 2. Inhibition of Rb phosphorylation by compound 4 and SU9516 in MDA-MB231 cells. Proteins were separated on SDS page, electroblotted to nitrocellulose, and hybridized with antibodies against phoshorylated Rb (T821, left panel and S780, right panel). Blots were subsequently hybridized with antibodies against total Rb and actin (used as loading control). C, control, untreated cells; SU, cells treated with SU9516 (10 μM); 4, cells treated with compound 4 (4 μM).
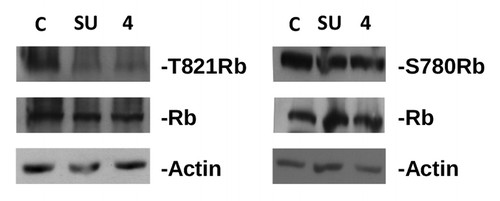
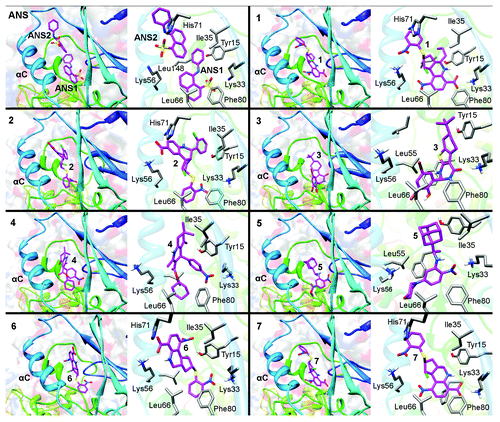
Figure 3. Predicted binding modes of compounds 1–7 in the allosteric pocket of CDK2, highlighting (left) the location of each compound in the overall fold, (right) the close up of the allosteric site with selected residues interacting with the ligands. For comparative purposes, the first box shows the binding mode of the 2 ANS molecules present in the crystal structure of the binary complex (PDB code 1PXF).