Figures & data
Figure 1. T2AG3-dependant integration in humanized yeasts. (A) Wild-type (TLC1) or humanized yeasts (tlc1-h) are transformed with a non-replicative telomeric plasmid containing either T2AG3 or TG1–3. The rate of integration is determined by selection on G418 plates and normalized with the transformation efficiency obtained with a co-transformed replicative plasmid. (B) Humanized yeasts carrying tlc1-h template were transformed with a plasmid expressing the wild-type TLC1. A plasmid shuffle allows the strain to keep only the TLC1 or both the TLC1 and tlc1-h templates. (C) Genomic DNA was digested by XhoI and 1 kb TRF (telomere restriction fragments) are released. Southern blot was performed and DNA was revealed with 3 different probes: T2AG3, TG1–3 and Y’ (subtelomeric fragment, DNA loading control). Hybridization intensities are measured with Imagequant shareware. T2AG3 and TG1–3 intensities are normalized according to the Y’ intensity corresponding to the total DNA quantity. (D) Clones were transformed with the human-type telomeric plasmid. The rate of integration is determined by selection on G418 plates and normalized with the transformation efficiency obtained with a replicative plasmid co-transformed.
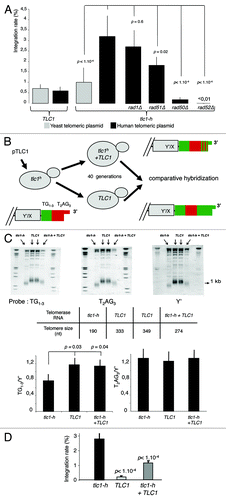
Figure 2. Recombinogenic properties of TRF2 in humanized yeasts. (A) In humanized yeasts, the VII-L distal end was replaced by the reporter gene HIS3. TRF2 is fused to a transcription activation domain (AD), and expressed in the humanized strain carrying the reporter gene. The humanized yeast strain (SD-URA) is replica-plated on SD-URA-HIS + 3-AT to monitor the expression of HIS3. (B) Scheme and western blots showing the structure and the inducibility of the TRF2, TRF2∆B, TRF2∆M, TRF2∆B∆M, or TRF1 genes. Glc, glucose; G/R, galactose. (C) The humanized yeasts expressing TRF2, TRF2∆B, TRF2∆M, TRF2∆B∆M, or TRF1 were transformed with the human-type telomeric plasmid. The rate of integration is determined by selection on G418 plates and normalized with the transformation efficiency obtained with a replicative plasmid co-transformed. (D) Clones coming from the humanized yeasts expressing TRF2 or TRF2∆B and having integrated the plasmid were restreaked 1 (R1), 2 (R2), 3 (R3), or 4 (R4) times on YPD and after restreaked on selective G418 medium. The rate of excision is determined by selection on G418 plates of clones that does not grown anymore. (E) In gel assay to determine the quantity of 3′ overhang in humanized yeasts containing various forms of human shelterin proteins.
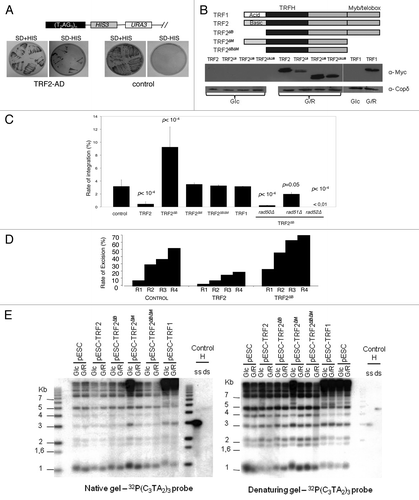
Figure 3. TRF2ΔB reduced the telomere length by trimming the leading strand telomere. (A) The genomic DNA from infected superHT1080 cells expressing an empty vector or TRF2ΔB was purified, digested and then separated on the same gel before to be transferred on a membrane. (T2AG3)n probe, specific to telomeric DNA, was used. The observed signal was analyzed to measure telomere length. Position of the mean telomere length is indicated by a white bar. Molecular weights (kb) are indicated at the bottom of the gel. (B) Preferential deletion of telomeric leading strand detected by qCO-FISH. Metaphase spreads of infected superHT1080 cells, expressing an empty vector or TRF2ΔB and incubated with BrdU/BrdC, were treated to remove the newly synthesized DNA strand. The remaining telomeric DNA strands were detected with PNA-C-Cy3 and LNA-G-FITC probes. Each telomeric signal was quantified as a telomeric intensity value (unit arbitrary, u.a.) and the mean telomeric intensity value was calculated for each strand (± s.e.m) and represented in graphic. Statistical analyses were done using the Wilcoxon rank-sum test. Ns is for no significant and *** is extremely significant with the P value < 0.001.
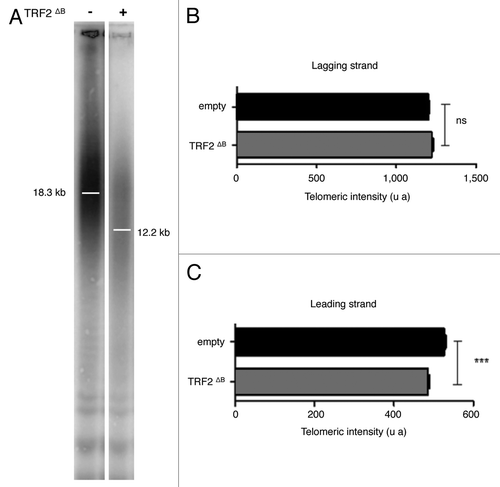
Figure 4. MUS81, GEN1, and SLX4 are required for TRF2∆B-mediated shortening in human cells. (A) At the top, total RNA from infected superHT1080 cells, expressing the pLKO.1 vector shMUS81, shGEN1, shSLX4, or sh non target, was purified and analyzed by RT-qPCR analysis. The graph represents the percent of mRNA inhibition for each MUS81, GEN1, and SLX4 mRNAs. At the bottom, proteins from infected superHT1080 cells were extracted and analyzed by western blot. (B) TRF length analysis of superHT1080 cells infected first with lentivirus expressing shMUS81, shGEN1, shSLX4, or sh non target and then with an empty control or TRF2 ΔB lentivirus. The different conditions are indicated above the gel. The genomic DNA was purified; equal amounts were digested by RsaI and HinfI restriction enzymes, and then separated on the same gel before to be transferred on a membrane. (T2AG3)n probe was used. The bars on the gel image indicate the position of the telomeric average size. On the left, the numbers indicate the size corresponding to the ladder (in kb). (C) Q-FISH analysis to measure the telomere length distribution of superHT1080 cells infected with TRF2 ΔB lentivirus. Both telomeric DNA strands were detected with PNA-C-Cy3 probe. Mean telomere lengths given as mean ± s.e.m. Statistical analyses were done using the Wilcoxon rank–sum test (***P < 0.001 = highly significant).
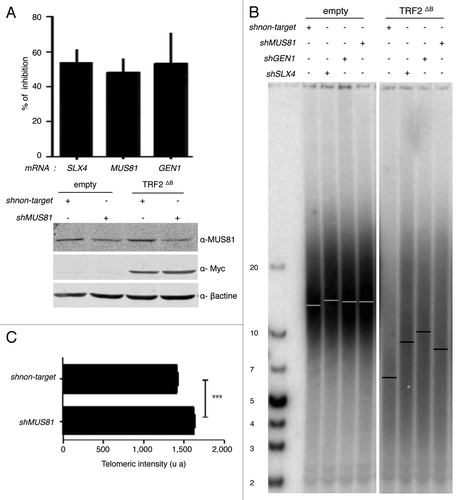
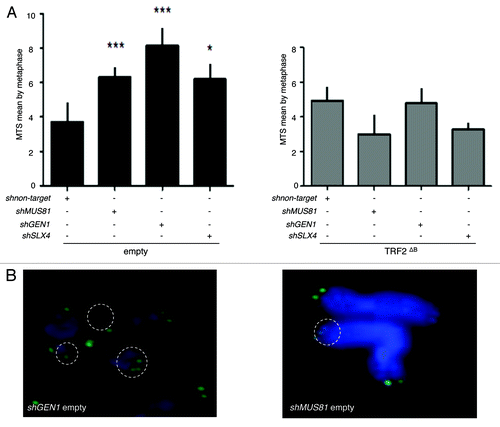
Figure 5. SLX4, MUS81, and GEN1 are required for telomere stability. (A) Quantification of the average number of multi-telomeric signals (MTS) per superHT1080 metaphase spreads (mean ± s.e.m.). Statistical tests: ***P < 0.001 = highly significant; **P value from 0.001 to 0.01 = very significant; *P value between 0.01 and 0.05 = significant; ns P value > 0.05 = not significant. (B) Images of superHT1080 metaphase spreads showing examples of MTS (white circles). Green, PNA-C-Cy3 probe; blue, DAPI.