Figures & data
Figure 1 IRA deletion strains are sensitive to cdc13-1-induced arrest despite an intact DNA damage checkpoint. (A) 10-fold serial dilutions were spotted to YPD agar and incubated at the indicated temperature for three days. (B) Log phase cultures were sonicated, spread onto 30° YPD agar and incubated at 30° for 8 hours. 100 colonies were scored for each strain. Values are mean ± standard deviation of three independent assays. (C) Cultures were arrested with α-factor at 21°, then raised to 32° to inactivate cdc13-1, then released from the α-factor block at 32°. Samples were sonicated, counted, serially diluted, and plated to 21° YPD agar. Viability is calculated as (CFU/mL)/(Total cells per mL) normalized to α-factor samples. Values are mean ± standard deviation of three assays. **p < 0.01, ***p < 0.001 by student's t-test. (D) YPD plates from (C) were inspected 18 hours after returning to 21°. Between 50 and 60 colonies were scored for each strain. Values are mean ± standard deviation of three assays.
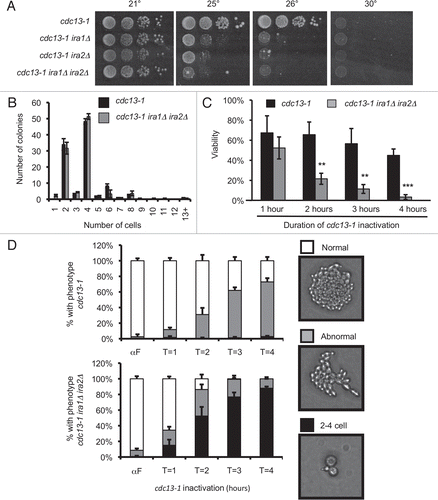
Figure 2 IRA deletion cannot restrain mitosis in the absence of an intact checkpoint. Cultures were treated as in . Values are mean ± standard deviation of three independent assays. Graphs represent microcolony assays for (A) chk1Δ, (B) rad53-21 and (C) pds1Δ backgrounds.
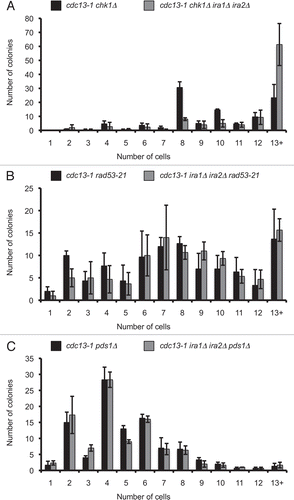
Figure 3 IRA deletion strains dephosphorylate Rad53 under conditions that delay checkpoint recovery. Cultures were synchronized with α-factor in YPD pH 3.9, raised to 32° for 75 minutes, and α-factor was removed by washing cells with 32° YPD pH 6.2. Cultures were held at 32° for four hours, then returned to 21°. Protein samples were harvested by TCA precipitation and analyzed by western analysis for Rad53 phosphorylation (A) and cells were fixed and analyzed for nuclear division after 0–180 minutes of recovery (C). In (B), samples were run adjacent to extracts from cultures treated with 10 µg/mL nocodazole for 4 hours at 21°. >90% of cells were large-budded at the time of harvest.
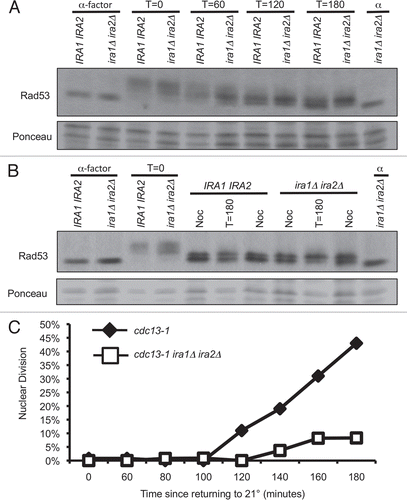
Figure 4 cdc13-1 ira1Δ ira2Δ has delayed recovery kinetics. α-factor arrested cultures were raised to 32° and released from the G1 arrest by micromanipulation onto 32° YPD agar. After three hours at 32°, plates were returned to 21° (T = 0 hours recovery). Colonies were scored at the indicated times and classified as visible to the naked eye (Recovered), microscopically visible with more than 2 cells (Microscopic) or as single large-budded cells (2-cell). Values are mean ± standard error of three assays. (A) cdc13-1, (B) cdc13-1 ira1Δ ira2Δ.
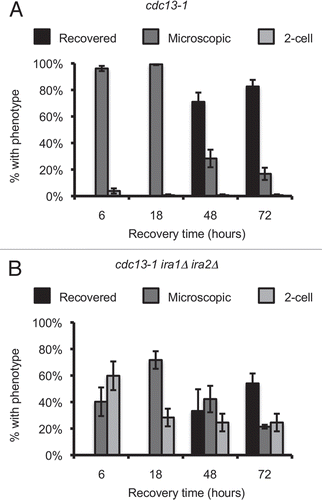
Figure 5 Single-cell recovery assays reveal signaling elements required for the ira1Δ ira2Δ recovery defect. (A and B) Strains were treated as in . Colonies were scored after 72 hours of recovery. (C) Strains were treated as in except cultures were in synthetic complete media lacking leucine (C-Leu) to maintain CDC20 plasmids. Values are mean ± standard error of at least three assays.
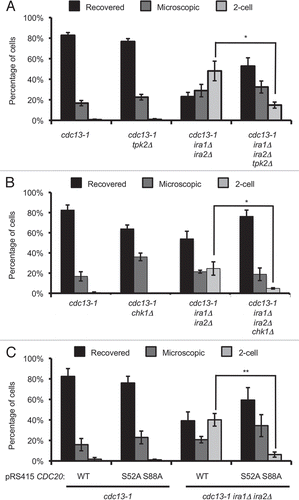
Table 1 Strains and constructs used in this study