Figures & data
Figure 1 Effects of miR-31 on tumor cell migration and metastasis are cell type and tissue dependent.
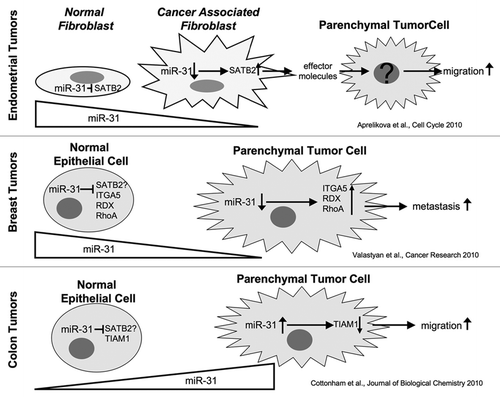
Figure 1 Model for mutually exclusive formation of RNA polymerase I, II and III transcription factories within the yeast rDNA. Interactions bridging the IGS1-IGS2 (1) and the 25S rDNA-IGS1 (2) sequences form part of an RNA polymerase II transcription factory promoting transcription from cryptic RNA polymerase II promoters. These interactions isolate active 5S rDNA in yeast. The 25S - IGS1 interaction is mutually exclusive with the intra-35S (18-25S) rDNA loop (3) that positions IGS1 and IGS2 sequences for rapid re-initiation of RNA polymerase I and requires Rep1p binding and blockage at the replication fork binding site. For simplicity figures are depicted in two-dimensions, in reality the structures would form rosettes that merge the two halves (1, 2) of the polymerase II factories. The rosette structure would also merge the RNA polymerase I factories (3, 4). Proximity-based ligation methods are unable to differentiate interactions occurring within a repeat (3) from those occurring between repeats (4), yet they are structurally different. Int. seq.: intervening sequence that could include no, one or several rDNA repeats.
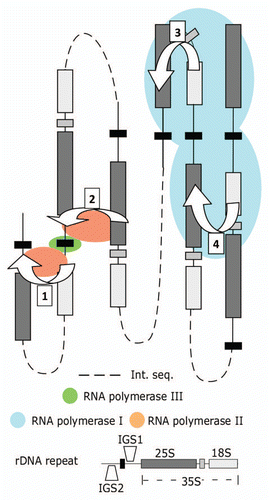
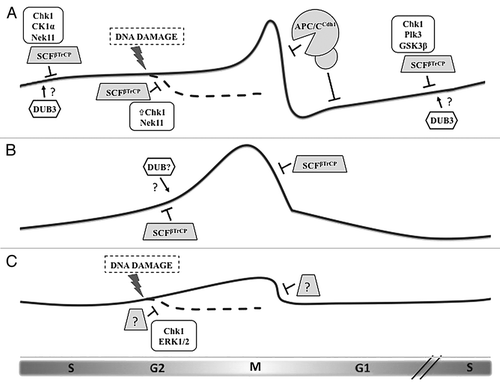
Figure 1 Regulation of the three CDC25 paralogs during the cell cycle by ubiquitin-mediated degradation. (A) Cdc25A protein levels begin to rise in late G1 and remain modest during S phase and early G2 by the action of the ubiquitin ligase SCFβTrCP, whose recognition of Cdc25A depends upon phosphorylation by a number of kinases. DNA damage induction of Chk1 activity increases the priming phosphorylation of Cdc25A, leading to greater βTrCP-induced degradation. At the end of mitosis and through G1, Cdc25A is eliminated via the APC/CCdh1 ubiquitin ligase. Dub3 has been shown to stabilize Cdc25A by deubiquitylation, although the precise timing and conditions for its activity remain unknown. (B) Cdc25B levels are also moderated by βTrCP, but in a phosphorylation-independent manner. Rapid destruction of Cdc25B at the metaphase-anaphase transition is mediated by βTrCP. Since the degron of Cdc25B mimics constitutive phosphorylation, we hypothesize that another mechanism counteracts βTrCP-mediated ubiquitylation during its rise in G2, possibly the activity of a deubiquitylating enzyme (DUB). (C) Cdc25C is the least-studied protein of the family, and its regulation is determined mostly by phosphorylation events that regulate its phosphatase activity and localization. Evidence exists that Cdc25C is ubiquitylated and degraded following certain forms of G2 arrest, but, although this event requires Chk1 and ERK1/2, the contributing ubiquitin ligase is unknown.