Figures & data
Figure 1 kin-19/CKIα expression during C. elegans larval development. kin-19::GFP translational reporter expression in the (A) epidermal seam cells (SC), (B) vulval cells (V) and neuronal cells (N) of the ventral nerve cord in L4 stage wildtype animals. Images taken at 430×. (C) Expression levels of kin-19 transcript, relative to ama-1 transcript, during larval stages in wildtype C. elegans grown at 25°C.
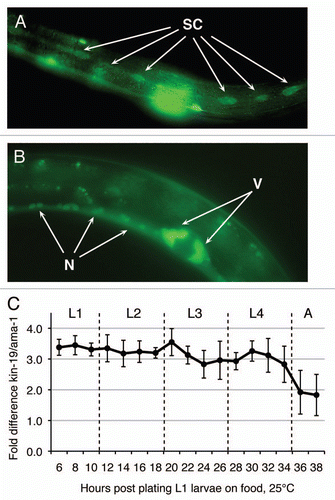
Figure 2 Anomalous terminal seam cell numbers result from RNAi knockdown of kin-9, pop-1 or wrm-1. (A–D) Young adult or L4 wIs78 worms, in which GFP expression marks the seam cell nuclei (SCM::GFP) and adherens junctions (ajm-1::gfp/MH27::GFP). Images are composites of 3 to 4 photographs of the same animal in each case, taken at a magnification of 200×. (A) Young adult wIs78 animal on mock RNAi showing 14 seam cell nuclei within the continuous syncytium of the fused seam cells. (B) Young adult wIs78 animal on kin-19(RNAi) showing 27 seam cell nuclei. (C) Young adult wIs78 animal on pop-1(RNAi) showing 48 seam cell nuclei. (D) Mid L4 stage wIs78 animal on wrm-1(RNAi) showing 9 seam cell nuclei (SC) within the discontinuous seam cell syncytium. Also visible in the plane of the seam cells are the nuclei of cells (Hyp) that have not fused with the seam cell syncytium.
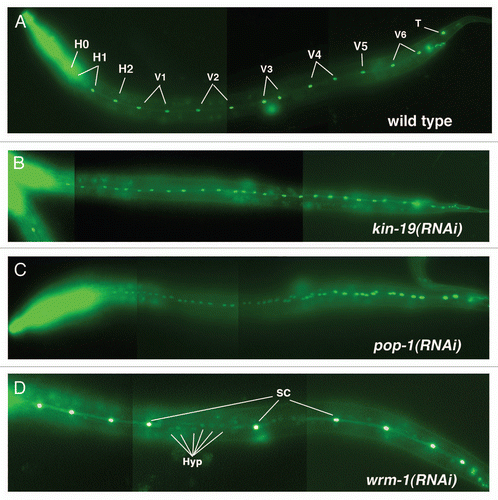
Figure 3 (A–H) Anomalous seam cell numbers result from RNAi knockdown of kin-19 and Wnt signaling pathway genes. For each RNAi condition, n = 45–50 animals were observed for each time point, and data points are averages of three trials. Error bars indicate standard deviation. (A) wIs78 animals on mock RNAi, and RNAi of bar-1, hmp-2 or sys-1 showed wildtype seam cell numbers and pattern of division. (B) RNAi knockdown of kin-19, pop-1 or apr-1 resulted in seam cell hyperplasia starting in the early L4 stage and persisting into the adult stage. (C) RNAi knockdown of wrm-1, lit-1 or mom-4 resulted in seam cell hypoplasia, starting during the late L2 stage and persisting into the adult stage. (D) RNAi knockdown of the heterochronic genes kin-20, lin-42 or lin-41 resulted in the normal terminal seam cell number, but fewer than normal seam cells underwent division in the L4 stage. hbl-1(RNAi) produced a similar seam cell pattern of division and terminal seam cell number (not shown).
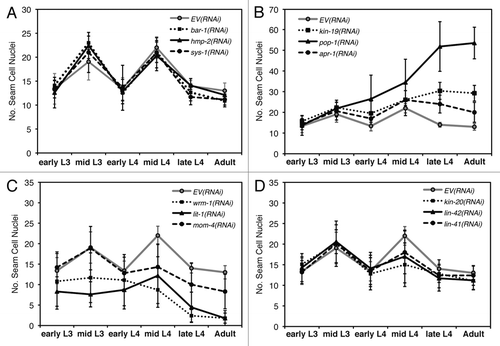
Figure 4 Post-embryonic lineages of epidermal seam cells V1–4 and V6 on RNAi knockdown of wnt signaling pathway genes and/or heterochronic genes. H: hypodermal cell, SC: seam cell, SC boxed in black indicates terminally differentiated seam cells that have fused into a syncytium. L1–L4: larval stages 1 to 4, A: adult. Numbers in parentheses at the bottom of each lineage indicate the number of terminal seams cells within the lineage shown (first number), and the range of terminal seam cells that results if all V-seam cells undergo the lineage shown (second number). (A) Normal pattern of seam cell (V1–4, 6) division and differentiation during larval development. (2/14) Each of the V1–4, V6 lineages produce 2 terminal seam cells, and the total number of visible terminal seam cells is 14. (B–D) Models of misspecification of V seam cell lineages in worms on (B) kin-19(RNAi), (C) pop-1(RNAi) and (D) wrm-1(RNAi) or lit-1(RNAi). (E–H) Models of misspecification of V seam cell lineages in worms on (E) RNAi of precocious heterochronic genes (PHG-kin-20, lin-42, lin-41 or hbl-1), and (F–H) simultaneous RNAi of a PHG and a wnt signaling pathway gene—(F) kin-19, (G) pop-1, (H) wrm-1 or lit-1.
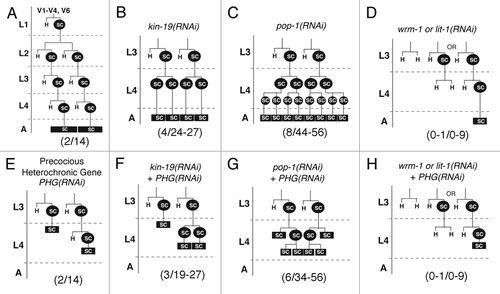
Figure 5 Effect of genetic interaction between kin-19 and Wnt signaling pathway genes on terminal seam cell number, a measure of asymmetric division. (A and B) The number of terminal seam cell nuclei was observed in wIs78 animals fed on simultaneous RNAi of kin-19 and the indicated Wnt signaling pathway gene, except for the following in (A): wrm-1(ne1982ts);wIs78 or lit-1(ne1991ts);wIs78 mutant animals were grown at and subjected to RNAi of the indicated Wnt signaling pathway gene at 25°C. Results shown are averages of three trials carried out with n=50–65 animals for each RNAi condition. Error bars indicate standard deviation. (**) P< 0.05 and (*) P<0.1, indicates statistically significant suppression of kin-19/EV(RNAi) phenotype, determined using students t-test. The dashed line indicates the average terminal seam cell number on kin-19(RNAi). The dotted line indicates the wildtype terminal seam cell number.
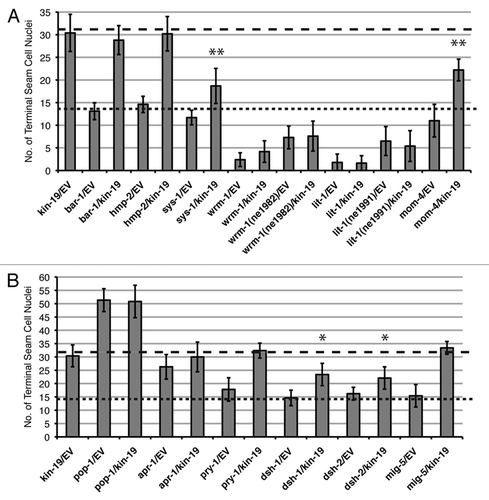
Figure 6 Effect of genetic interaction between kin-19 and heterochronic pathway genes on terminal seam cell number, a measure of asymmetric division. The number of terminal seam cell nuclei was observed in wIs78 animals fed on simultaneous RNAi of kin-19 and the indicated heterochronic pathway gene. Results shown are averages of three trials carried out with n = 50–65 animals for each RNAi condition. Error bars indicate standard deviation. **p < 0.05 and *p < 0.1, indicates statistically significant suppression of kin-19/EV(RNAi) phenotype, determined using students t-test.
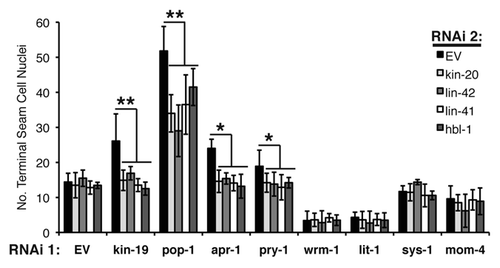
Figure 7 Effect of genetic interaction between kin-19 and heterochronic pathway genes on alae formation, a measure of terminal differentiation. The extent of alae expression was observed in young adult wIs78 (A) or let-7(n2853) (B) worms on RNAi of the indicated Wnt signaling pathways genes. The extent of alae expression was observed in early L4 stage lin-42(n1089) (C), lin-41(ma104) (D) hbl-1(mg285) (E) and wIs78 (F) animals on RNAi of the indicated Wnt signaling pathways genes or double RNAi with kin-20 (F). Results shown are the average of three trials of each experiment carried out with n = 30–40 animals for each RNAi condition. Error bars indicate standard deviation. **p < 0.05 and *p < 0.1, indicates statistically significant suppression compared to EV(RNAi), determined using students t-test.
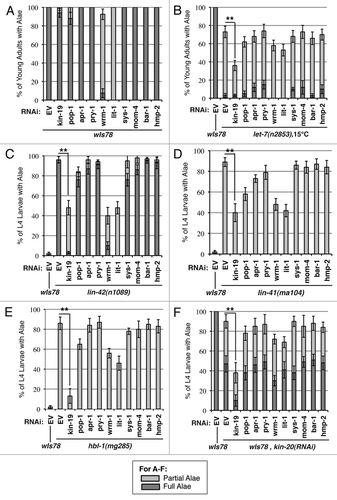
Figure 8 Effect of genetic interaction between kin-19 and heterochronic pathway genes on seam cell fusion, a measure of terminal differentiation. (A) Seam cell fusion in early and mid-L4 animals of the following strains: wIs78, lin-42(n1089), lin-42(ma104), hbl-1(mg285) on RNAi of the indicated Wnt asymmetry pathway genes. (B) Seam cell fusion in early and mid-L4 wIs78 animals on simultaneous RNAi of kin-20 and the indicated Wnt asymmetry pathway gene. For (B), the dashed line indicates the average number of L4 stage wIs78 animals on kin-20(RNAi) showing precocious seam cell fusion. Error bars indicate standard deviation. **p < 0.05 and *p < 0.1, indicates statistically significant suppression compared to EV(RNAi), determined using students t-test.
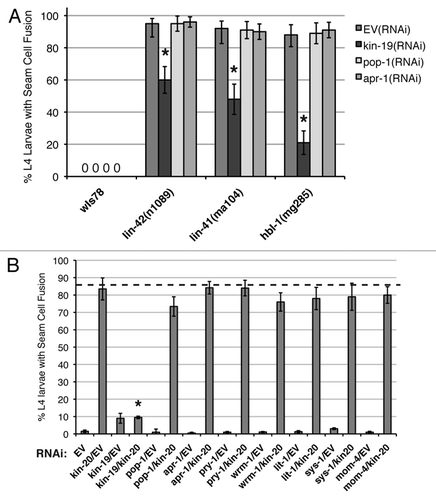
Figure 9 Models of kin-19 function in C. elegans seam stem cells. (A) A simplified model of differential fate specification in daughter cells mediated by the Wnt/β-catenin asymmetry pathway, and the possible roles of KIN-19 in this pathway. FZ: Frizzled, Wnt receptor, PM: plasma membrane, NM: nuclear membrane, P: phosphorylation. Pathway components that do not appear to function in the V seam cell asymmetry pathway (Wnt, FZ) are shown in grey. Indirect and speculative interactions are indicated by dashed, grey lines. (B) The relationship between kin-19 and the late larval heterochronic genes in regulating seam cell terminal differentiation. kin-19 likely functions downstream of and in parallel to the heterochronic genes to act as a positive regulator of terminal differentiation.
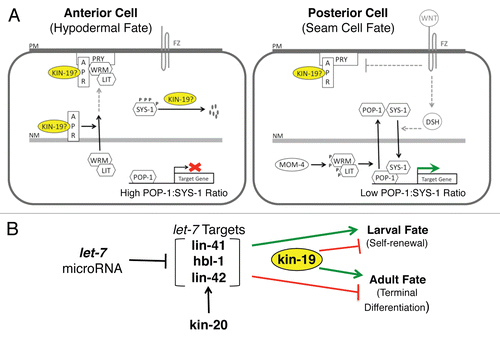
Table 1 Terminal seam cell number resulting from genetic interaction between Wnt singaling pathway genes