Figures & data
Figure 1. Pannexins are N-glycosylated but not O-glycosylated as defined by sensitivity to enzymatic digestion. Upon treatment of total protein lysates from HEK-293T cells ectopically expressing Panx1, 2, or 3 with PNGase F, a significant gel mobility shift in western blots was observed for Panx1 (A) and Panx3 (C) with a moderate change in mobility for Panx2 (B), indicating that all 3 pannexins are N-glycosylated. Sequential treatments with sialidase A and O-glycanase resulted in a change in the banding pattern of Fetuin in a Sypro-stained gel, used as a positive control (D), but did not generate visible banding shifts in any of the pannexin labeled blots (A, B, and C). The position of molecular weight standards are shown in kDa.
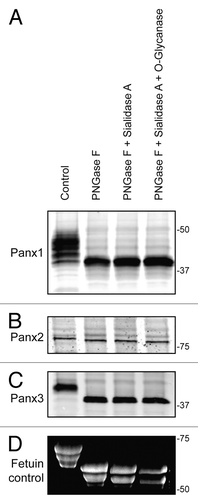
Figure 2. Panx1 and Panx2 are substrates for caspase cleavage. In vitro caspase 3 and caspase 7 cleavage of Panx1, -2, and -3, ectopically expressed in HEK-293T cells. Panx-CT antibodies were used for pull down (IP) of all 3 pannexins, treated with: buffer only, exogenous caspase 3 or 7 (500 nM), or both caspase 3 (500 nM) and the pan-caspase inhibitor Z-VAD-OMe-FMK (50 µM). After immunoblotting (IB) with Panx-CT antibodies, the absence of protein bands indicated caspase cleavage of Panx1 (A) and Panx2 (B) as compared with controls. The presence of Panx3 protein bands despite the addition of activated exogenous caspase suggested that Panx3 is not a substrate for caspase-dependent cleavage in vitro. The IgG (50 kDa) antibody heavy chain is found in all 3 blots as expected. Cleaved protein bands detected at ~32 kDa for Panx1-GFP (D) and ~63 kDa for Panx2-GFP (E) suggested that the caspase cleavage sites are located in the C-terminal domains of both proteins. Cleavage is inhibited in the presence of the pan-caspase inhibitor Z-VAD. Protein sizes are noted in kDa.
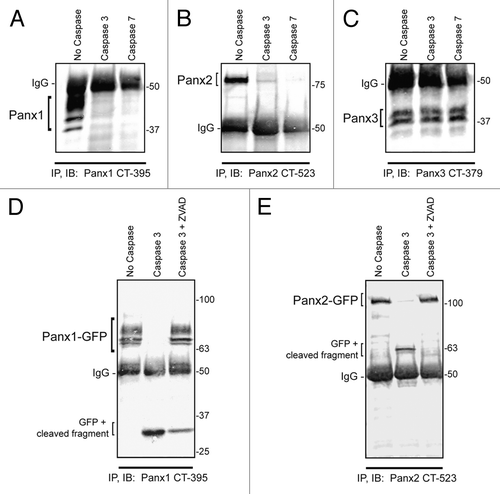
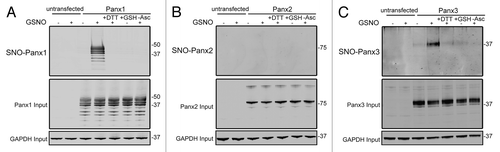
Figure 3. Panx1 and Panx3 are post-translationally modified by S-nitrosylation. Western blots for Panx1 (A), Panx2 (B), and Panx3 (C) following an assay for detection of S-nitrosylated proteins. HEK-293T cells ectopically expressing murine Panx1, Panx2, or Panx3 were treated with 100 µM GSNO to facilitate S-nitrosylation and samples were processed by the Biotin Switch assay. For controls, Panx expressing cells were stimulated with 100 µM reduced GSH, the backbone molecule in GSNO that lacks the capacity to release NO, or with the reducing agent DTT (1mM) following GSNO treatment to reduce S-nitrosylated protein thiols. As a negative control, ascorbate was omitted from the reducing step to prevent reduction of S-nitrosylated cysteines and subsequent biotinylation. GAPDH was used as input controls. Protein sizes are noted in kDa.