Figures & data
Figure 1 Null mutation of the BqsS-BqsR two-component system results in enhanced biofilm formation in P. aeruginosa strain mPAO1. (A) Genetic organization and domain structures of the sensor kinase BqsS and the response regulator BqsR. Gene orientation is indicated by arrow. Domain structure prediction was done using the SMART program (http://smart.emblheidelberg.de/). Symbol: REC, cheY-homologous receiver domain: reg_C: response regulator receiver domain; SP, signal peptide; HAMP, histidine kinases, adenylyl cyclases, methyl binding proteins, phosphatases domain; HisKA, phospho-acceptor domain; HATPase, histidine kinase-like ATPase domain. (B) Visualization of biofilm formation on the walls of the polystyrene tubes by crystal violet staining. (C) Quantification of biofilm formation. The data shown are the means of triplicates and the standard deviation (SD) is shown by error bar. The following bacterial strains were used in this experiment: mPAO1 (mP), the bqsS deletion mutant ΔbqsS (ΔS), the bqsR mutant ΔbqsR (ΔR), and the complemented strains ΔbqsS(bqsS) [ΔS(S)], ΔbqsS(bqsR) [ΔS(R)], ΔbqsR(bqsR) [ΔR(R)] and ΔbqsR(bqsS) [ΔR(S)].
![Figure 1 Null mutation of the BqsS-BqsR two-component system results in enhanced biofilm formation in P. aeruginosa strain mPAO1. (A) Genetic organization and domain structures of the sensor kinase BqsS and the response regulator BqsR. Gene orientation is indicated by arrow. Domain structure prediction was done using the SMART program (http://smart.emblheidelberg.de/). Symbol: REC, cheY-homologous receiver domain: reg_C: response regulator receiver domain; SP, signal peptide; HAMP, histidine kinases, adenylyl cyclases, methyl binding proteins, phosphatases domain; HisKA, phospho-acceptor domain; HATPase, histidine kinase-like ATPase domain. (B) Visualization of biofilm formation on the walls of the polystyrene tubes by crystal violet staining. (C) Quantification of biofilm formation. The data shown are the means of triplicates and the standard deviation (SD) is shown by error bar. The following bacterial strains were used in this experiment: mPAO1 (mP), the bqsS deletion mutant ΔbqsS (ΔS), the bqsR mutant ΔbqsR (ΔR), and the complemented strains ΔbqsS(bqsS) [ΔS(S)], ΔbqsS(bqsR) [ΔS(R)], ΔbqsR(bqsR) [ΔR(R)] and ΔbqsR(bqsS) [ΔR(S)].](/cms/asset/076cc389-a93a-4a32-bb72-5f50c1e98081/kcib_a_10906717_f0001.gif)
Figure 2 Time course assay of biofilm development. (A) Quantification of biofilm formation at different time points. P. aeruginosa strains were grown in LB medium in 14-ml polystyrene tubes and the amounts of biofilm mass at different time points as indicated were quantified. Assays were performed in triplicates and the data were the mean values ± SD. (B) Bioscreen analysis of the bacterial growth. The data were the mean values of 5 replicates. The following bacterial strains were used in this experiment: mPAO1 (mP), mutant ΔbqsS (ΔS), mutant ΔbqsR (ΔR), and the complemented strains ΔbqsS(bqsS) [ΔS(S)], and ΔbqsR(bqsR) [ΔR(R)].
![Figure 2 Time course assay of biofilm development. (A) Quantification of biofilm formation at different time points. P. aeruginosa strains were grown in LB medium in 14-ml polystyrene tubes and the amounts of biofilm mass at different time points as indicated were quantified. Assays were performed in triplicates and the data were the mean values ± SD. (B) Bioscreen analysis of the bacterial growth. The data were the mean values of 5 replicates. The following bacterial strains were used in this experiment: mPAO1 (mP), mutant ΔbqsS (ΔS), mutant ΔbqsR (ΔR), and the complemented strains ΔbqsS(bqsS) [ΔS(S)], and ΔbqsR(bqsR) [ΔR(R)].](/cms/asset/4fd77939-4dc9-4390-b1b9-d712ba66aa9d/kcib_a_10906717_f0002.gif)
Figure 3 Effect of co-culture and wild-type supernatants on biofilm formation by mutant ΔbqsS. (A) Biofilm formation by co-cultured bacteria. The mutant ΔbqsS (ΔS) was mixed with wild-type mPAO1 (mP) in 2:1 or 1:1 ratio and biofilms were quantified after incubation for 24 h. (B) Biofilms formed in conditioned medium. Wild-type mPAO1 and mutant ΔbqsS were cultured in conditioned medium which contained 12.5% or 25% of the supernatants of mPAO1 in LB broth. The data are the means of triplicates and SD is shown by error bar.
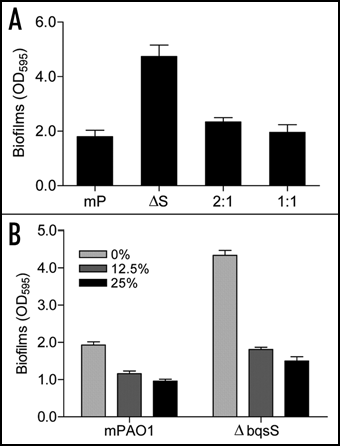
Figure 4 Effect of bqsS-bqsR mutation on rhamnolipid production and rhlA expression. (A) TLC plate assay of rhamnolipids production. P. aeruginosa strains described in were grown at 37°C for 24 h, and rhamnolipids were extracted from the supernatants for TLC analysis using standard rhamnolipids (Rh) as a positive control and the extracts from lasI and bqsS double mutant (IS) was used as negative control. Two predominant rhamnolipids, mono-rhamnolipids (mono-RL) and di-rhamnolipids (di-RL) were indicated by arrows. (B) The rhlA'-lacZ fusion gene expression assay. Different bacterial strains containing the prhlA-lacZ construct were grown in LB broth at 37°C to OD600 of 1.5 and the cells were then collected and assayed for β-galactosidase activity. The data were the means of triplicate with standard deviations. The bacterial strains used were indicated in .
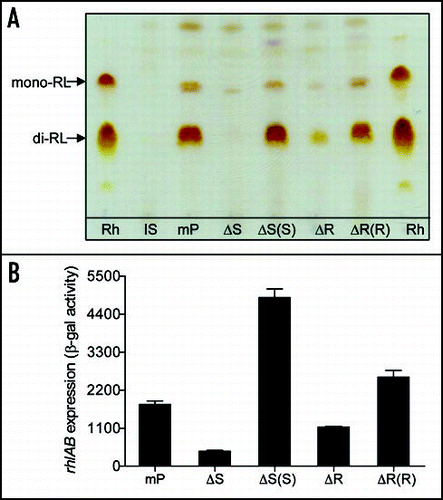
Figure 5 Effect of bqsS and bqsR deletion on QS signal production. (A) C4HSL production. C4HSL was extracted from the supernatants of P. aeruginosa strains and assayed by using indicator strain Chromobacterium violaceum CV026. Synthetic C4HSL was used as a positive control. Quantification was performed as describedCitation48,Citation50 and the data were presented as relative percentages to wild-type. (B) Pyocyanin production by P. aeruginosa strains as indicated. (C) TLC analysis of PQS (left) and anthranilate (right). PQS and anthranilate were extracted from supernatants of P. aeruginosa strains. The extracts were separated on TLC plates and the plates were visualized under UV (left) or natural light (right). Synthetic PQS (37 nmole) and anthranilate (25 nmole) were spotted separately as controls. (D) RT-PCR analysis of the transcript levels of pqsA and phnA. The proC gene, which is constitutively expressed, was amplified under the same conditions as an internal loading control.
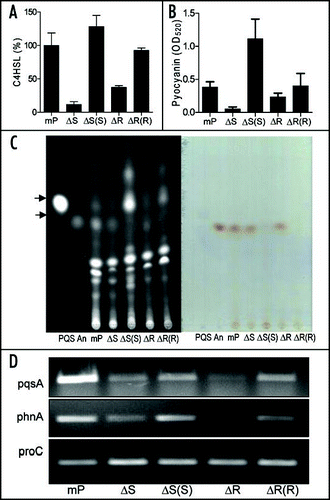
Figure 6 Effect of exogenous addition of rhmnolipids, C4HSL and PQS on biofilm formation by mutant ΔbqsS. P. aeruginosa strain mPAO1 (mP) was grown in LB medium as a control, and ΔbqsS (ΔS) was inoculated in the same medium with or without rhamnolipids (Rh, 50 µg/ml) or C4HSL (50 µM) or PQS (50 µM). The bacteria were grown for 6, 16, 24 and 32 h before quantification of biofilm formation. The data were the means of triplicates with standard deviation.
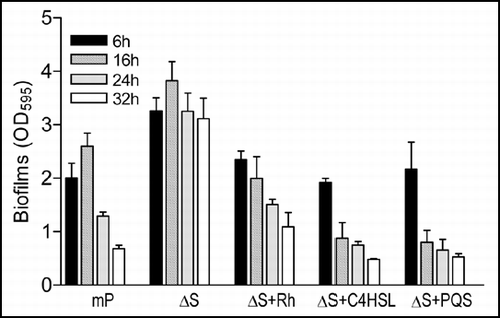
Table 1 Bacterial strains and plasmids used in this studyTable Footnotea