Figures & data
Figure 1 (At left) A model for CLV pathway signaling at the shoot meristem. Apically-generated CLV3 signal primarily activates CLV1 on the apical face of L3 stem cells within the shoot meristem. CLV1 in turn negatively regulates POL/PLL1 primarily on the apical portion of the cell. Upon periclinal division, the asymmetric distribution of POL/PLL1 drives apical/basal fates, including the activation of WUS in the basal daughter.
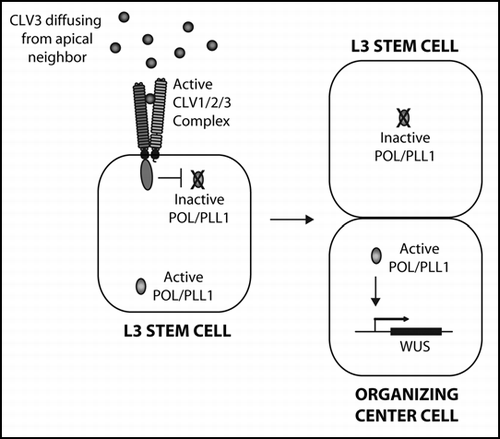
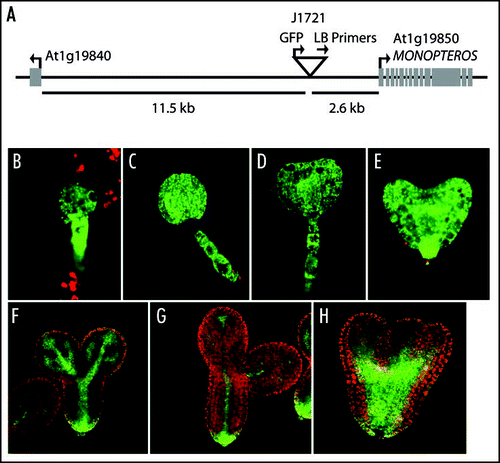
Figure 2 J1721 is located proximal to MP. (A). TAIL-PCRCitation20 products, obtained from J1721 seedlings using the degenerate primers, AGWGNAGWANCAWAGG and NGTCGASWGANAWGAA (where W = A or T, S = C or G, and N = A or C or G or T) in combination with the T-DNA left border primers: (Round 1) TTGATTTATAAGGGATTTTGCCGATTTCGG, (Round 2) AACTCTCTCAGGGCCAGGCG and (Round 3) CCACCCCAGTACATTAAAAACGTC, were sequenced to determine the insert location. The location of the T-DNA insertion is represented by an upside down triangle. Arrows indicate the directionality of genes and primers. The distance of the T-DNA from the start codons of the surrounding genes are as marked. (B–G). J1721 enhancer trap expression in green in various stages of wild-type sibling embryos from pol/pol pll1/+ parents. (H) J1721 expression in a pol pll1 embryo—note the broader and more intense signal.