Figures & data
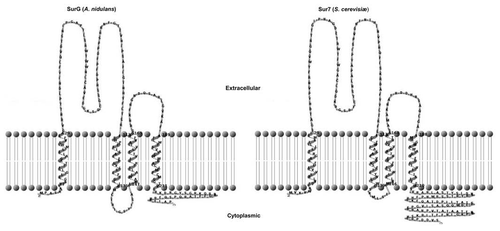
Figure 1 Phylogeny and Conservation of Pil-like proteins. (A) An unrooted maximum likelihood tree (for detail of methods see legend of Fig. 1 of ref. Citation3) showing the PilA and PilB clades of the Aspergilli. For reasons of clarity the systematic names of the autocalled genes are not shown in the tree, and are reported here. For the PilA clade: Afu: A. fumigatus, Afu6g07520; Nfi: Neosartoria fischeri, NFI_053190; Aor: A. oryzæ, AO090005001548; Afl: A. flavus, AFL2G_01442.2; Ate: A. terreus ATG _03673.1; Acl: A. clavatus ACL_081680; Ang: A. niger, An07g08890. For the PilB clade the same abbreviations are used and cognate systematic names are: Afu6g08320, NFI_053960, AO090003000094, AFL2G_02857.2, ATEG _05825.1, ACLA_082350, An11g01750. PilA and PilB denote the paralogues of A. nidulans (see text). The PilA and Lsp1 proteins of S. cerevisiae are also included in the tree. For reasons of space the aLRT branch support values are not shown within the PilA cluster, the lowest value being 0.74. (B) Eisosomal proteins in the Aspergilli. Comparison of the Pil1p, Lsp1 proteins of S. cerevisae and the PilA and PilB proteins of A. nidulans. Alignment carried out with MUSCLE, shading with Boxshade. Triangles, phosphorylation sites reported by Luo et al.Citation7 inverted triangles phosphorylation sites reported by Walther et al.Citation8
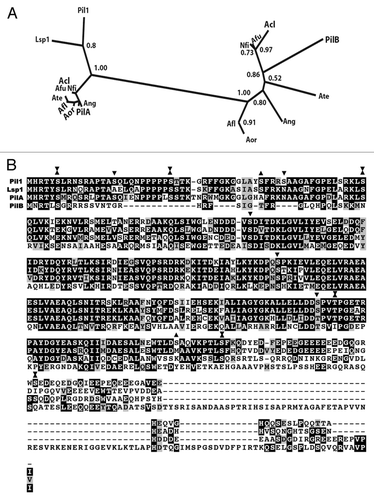
Figure 2 Proteins of the PilB clade have two putative coiled-coil domains. PilA and PilB proteins of the species of the Pezizomycotina included in the phylogeny of Figure 1 of reference Citation3 were independently aligned with T-Coffee. The alignments were scanned for coiled-coil domains with http://toolkit.tuebingen.mpg.de/pcoils. Size of window used by the programme: Green: 14 residues, blue: 21 residues, red: 28 residues. For the PilA clade, as a result of their high sequence identity, predictions for individual proteins do not differ from the prediction for the six aligned proteins. For the PilB clade, the sequence divergence leads to somewhat different individual predictions. The prediction for the coiled-coil domains of A. nidulans PilB is shown in the bottom panel.
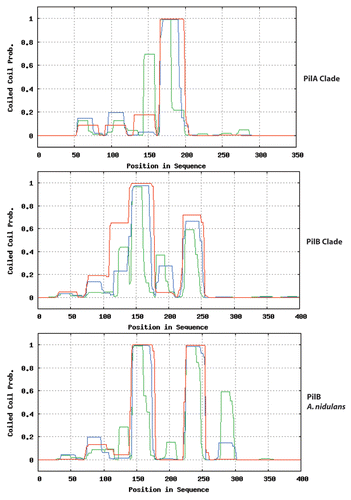
Figure 3 Predicted topology of SurG (A. nidulans) and Sur7 (S. cerevisiae). Graphic representation of the secondary structures of these proteins obtained with TMRP res2D (bioinformatics.biol.uoa.gr/TMRPres2D/index.jsp) based on the result from the TMHMM topology prediction tool (www.cbs.dtu.dk/services/TMHMM/).