Figures & data
Figure 1 Rab14 recruitment to chlamydial inclusions is a bacteria-driven process, independent of microtubules and Golgi integrity. HeLa cells infected with C. trachomatis serovar L2 (MOI 10) were fixed at 10 h p.i. Endogenous Rab14 was detected with rabbit polyclonal anti-Rab14 (1:100) followed by FITC-labeled goat anti-rabbit IgG (1:200) (green) in (A, C and D). Eukaryotic and bacterial DNA was evidenced by Hoescht staining (blue). (B) GFP-Rab14wt overexpressing cells (green) infected with C. trachomatis L2 (MOI 10) were incubated with 200 µg/ml Chloramphenicol (20 h) to inhibit bacterial protein synthesis. Cells were fixed at 24 h p.i. (C) Infected cells were treated with 1 µg/ml Brefeldin a (6 h) to disrupt Golgi apparatus. Mouse monoclonal anti-TGN 46 antibodies (1:200) followed by donkey anti-mouse Cy5-conjugated IgG (1:700) (red) were used to stain Golgi apparatus. (D) Infected cells were incubated with 20 µM Nocodazole (6 h) to depolymerize microtubules. Mouse anti-β-tubulin antibodies (1:200) followed by donkey anti-mouse Cy5-conjugated IgG (1:700) (red) were used to label microtubules. Insets show a magnification of the selected area of the cell. Bar, 10 µm. Endogenous Rab14 was clearly recruited to the inclusion as shown by rim-like green fluorescence staining around the entire periphery of the inclusion (A, C and D); whereas no recruitment was observed in Chloramphenicol-treated cells (B).
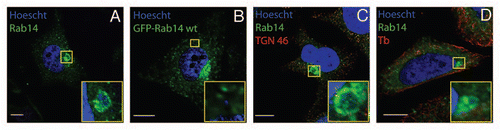
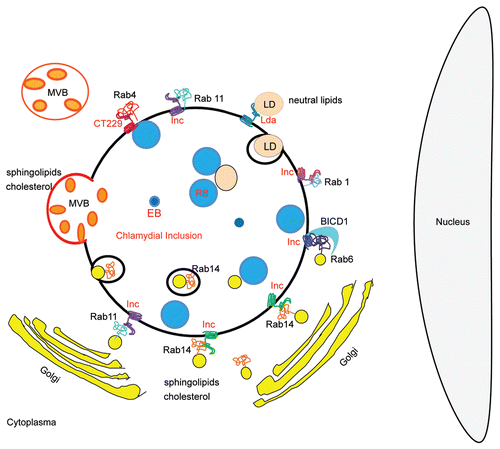
Figure 2 Schematic representation of lipid sources and Rab-controlled trafficking pathways usurp by Chlamydia trachomatis. RBs, Reticulate bodies are the replicative bacterial forms; EBs, Elementary bodies are the infective bacterial forms; Incs, Bacterial transmembrane proteins exposed at the inclusion membrane. Unidentified Incs are putative interacting factors with host proteins. CT229 is the Inc., from C. trachomatis that recruits Rab4; Rabs, Monomeric GTPases from host cells that control intracellular trafficking. Rab1, Rab4, Rab6, Rab11 and Rab14 have been found associated to C. trachomatis inclusion. BICD1 is a host Rab-6 interacting protein that is recruited to the chlamydial inclusion; MVBs, host multivesicular bodies, source of sphingolipids and cholesterol, are redirected to the chlamydial inclusion; LDs, Lipid droplets, host neutral lipids storage, are rerouted and translocated into the inclusion with the participation of chlamydial lipid droplet-associated proteins (Lda); Golgi-derived vesicles containing host endogenously-biosynthesized sphingolipids and cholesterol are targeted to the chlamydial inclusion. Note Golgi mini-stacks surrounding the inclusion. Chlamydia trachomatis intercepts Rab6-, Rab11- and Rab14-positive vesicles full of lipids, in transit from the Golgi apparatus to the plasma membrane. Rab14-labeled structures have been observed within inclusions at late-stages of chlamydial development.