Figures & data
Figure 1 Schematic diagram of the retromer complex and its role in endosome-to-Golgi retrieval. Retromer in S. cerevisiae is a stable heteropentamer comprising the cargo-selective VPS35-VPS26-VPS29 trimer and the SNX-BAR dimer of VPS5-VPS17. VPS29 links the two subcomplexes together.
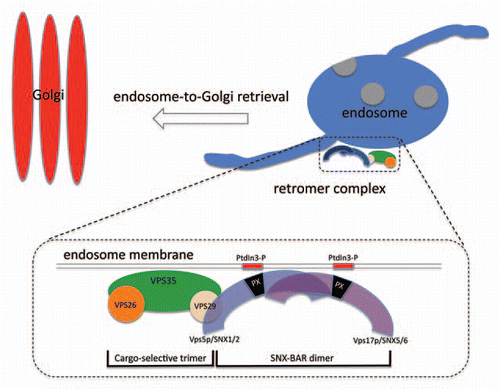
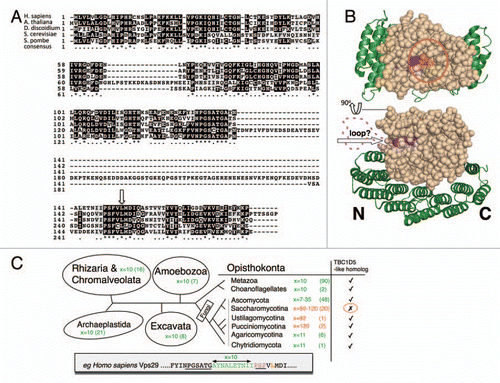
Figure 2 (A) Sequence alignment of VPS29 protein sequences from H. sapiens, A. thaliana, D. discoidium, S. pombe and S. cerevisiae. The conserved regions are shaded black. There is a large unconserved loop in the S. cerevisiae sequence that is not present in VPS29 from other species. The critical leucine involved in VPS29 interactions is highlighted with an arrow. (B) The additional loop is shown mapped onto the structure of mouse VPS29 (beige) in complex with the C-terminal domain of VPS35 (green) (based upon Hierro et al.Citation8). The loop lies close to the critical leucine residue (shown in orange) required for VPS29 to interact with Vps5 or TBC1D5. The purple shaded residues correspond to the conserved sequence P-S-F that is located just after the loop sequence. (C) VPS29 ancestral state. A survey of approximate distances (x, amino-acid residues) between the conserved PGSxTG and PSF motifs reveals that large sequence insertions are derived features in certain fungal clades (number of representatives considered in parentheses). Diagram adapted from Walker et al.Citation9 (see for further phylogenetic information). A TBC1D5 homolog was identified in all of the various eukaryotes except for Saccharomycotina.