Figures & data
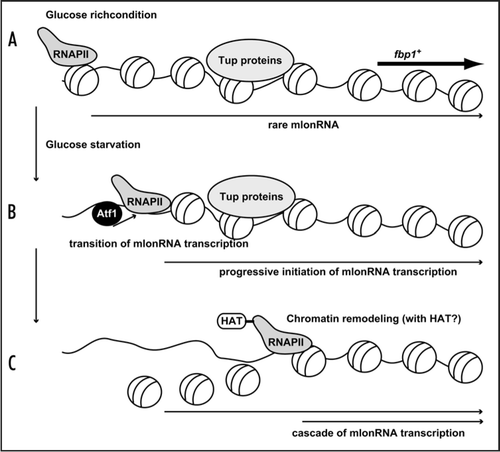
Figure 1 A model representing mlonRNA transcription disrupts chromatin array. (A) In glucose rich condition, rare mlonRNA is transcribed from a site far upstream from the authentic fbp1+ promoter, but does not initiate the robust activation of fbp1+ transcription at the promoter due to the Tup-dependent repressive chromatin structure. (B) Upon glucose starvation, Atf1 binds upper binding site (carrying CRE sequence). Atf1 activates progressive mlonRNA initiations, and this mlonRNA transcription overcomes the repressive role of the Tup proteins. (C) RNAPII traveling along upper fbp1+ region disrupts chromatin array possibly collaborating with HAT activity.