Figures & data
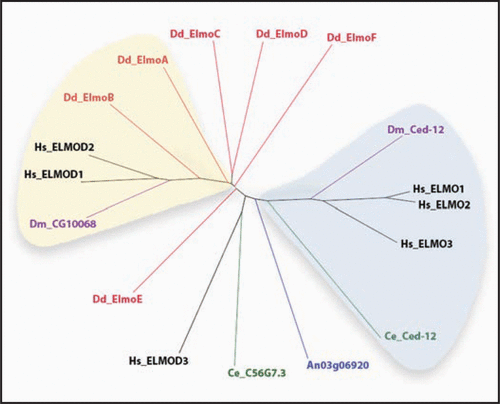
Figure 1 A relatedness tree of Elmo domains from proteins of five species is shown. The domain borders (indicated after each protein name below) were identified by a protein blast query of the full-length sequence. Species represented are Dictyostelium discoideum (Dd; red; elmoA: 364–551; elmoB: 106–266; elmoC: 363–547; elmoD: 288–456; elmoE (split domain): 477–566, 628–700; and elmoF (split domain): 259–350, 410–478), Homo sapiens (Hs; black; ELMO1: 300–482; ELMO2: 292–475; ELMO3: 175–356; ELMOD1: 115–304; ELMOD2: 112–272; ELMOD3: 151–314), Caenorhabditis elegans (Ce; green; C56G7.3: 123–293; Ced-12: 362–475), Drosophila melanogaster (Dm; purple; CG10068-PA: 121–290; Ced-12: 297–480) and Aspergillus niger (An; blue; AnCG5336: 220–410). Note that the Dd Elmo domains of elmoE and elmoF are split by poly-glutamine stretches, common in Dictyostelium proteins, which have been eliminated for the alignment. Highlighted in light blue are Elmo proteins reported to promote GEF activity. Highlighted in yellow are Elmo proteins speculated to promote GAP activity based on their proximity to the functionally characterized human ElmoD1 and D2 proteins and on preliminary results. Sequences were aligned with ClustalW2 (http://www.ebi.ac.uk/Tools/clustalw2/index.html). The resulting guide tree (dnd) was used to create a circular tree in the Java phylogenetic tree viewer Hypertree (http://kinase.com/tools/HyperTree.html; and ref. Citation24).