Figures & data
Figure 1. (A) Phylogeny of D. melanogaster (mel), its closest relatives (rel: D. simulans, D. sechellia, and D. mauritiana), and the outgroup species D. yakuba (yak). The Sperm dynein intermediate chain (Sdic) multigene family is only present in D. melanogaster and therefore it must have been originated after its lineage branched off from that leading to its closest relatives 5.4 mya.Citation36 (B) Details of the molecular organization of the Sdic multigene family at 19C1 of the X chromosome. Although four copies are annotated in FlyBase,Citation25 our computational analyses revealed the existence of additional haplotypes that denote the existence of further copies, in good agreement with earlier estimates (see main text). The presence of additional copies of the gene Sdic could conflict with models previously proposed for the generation of the multigene family.Citation6
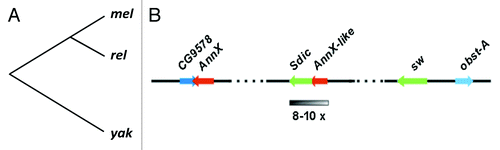
Figure 2. Molecular verification by RT-PCR of the proper expression of the sw transgene in testis of males with (A-) and without Sdic (B+). The lane of the underlined B+ corresponds to males carrying the endogenous copy of sw but not the transgene. H2O, negative control (no cDNA was added); L, ladder. Primers and methods are as described.Citation10
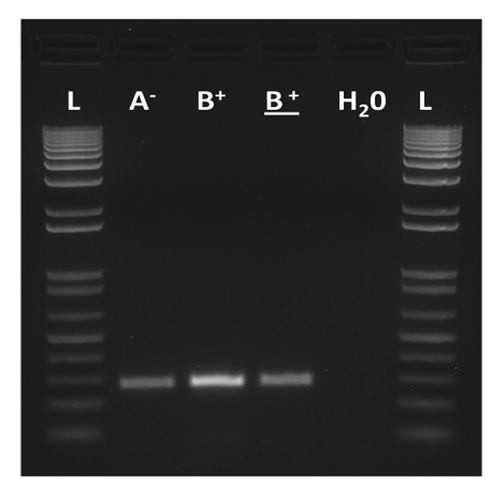
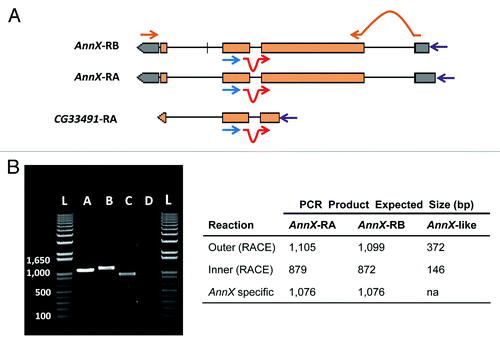
Figure 3. Functional test of the expression of AnnX-like genes. (a) Outline of the strategy followed. 5′ RACE experiments were performed to detect the presence of the putative transcripts corresponding to the AnnX-like genes in whole body and testis of 5-d-old males. The experiments were done following the instructions of the kit from Epicenter ExactSTART Eukaryotic mRNA 5′-&3′-RACE. Primers used: outer primer (blue; 5′GGATCCAAGTAGGGCTGTCA3′), located on the third exon of AnnX and the second exon of AnnX-like (only CG33491 is shown); inner primer (red; 5′GACTGGACGCACTCAACTATGG3′), located at the junction of the second and third exons; and a 5′ primer from the kit used (purple). The sequences of the outer and inner primers are complementary to those of the two AnnX transcripts and to that putatively transcribed from the AnnX-like genes according to FlyBase.Citation25AnnX transcript specific primers (brown) were used for RT-PCR experiments with the only purpose to test for the quality of the cDNA and the presence of genomic DNA. (B) Results for the expression profiling experiments in testis. Left, 5′ RACE products amplified from cDNAs of total RNA from males carrying the wild-type configuration for the Sdic multigene family (B+). Lanes: 1) PCR product using AnnX specific primers; 2) PCR product using the outer primer and the 5′ primer; 3) PCR product using the inner primer and the 5′ primer; 4) H2O, negative control (cDNA but no primers added); L) ladder. Right, summary table with the expected size of the PCR products for the two transcripts of AnnX and for the putative transcript of AnnX-like genes. Evidence for the presence of the two AnnX transcripts was detected but not for the putative transcript of AnnX-like genes. Experiments using total RNA from whole bodies as starting material shown identical results (not shown).