Figures & data
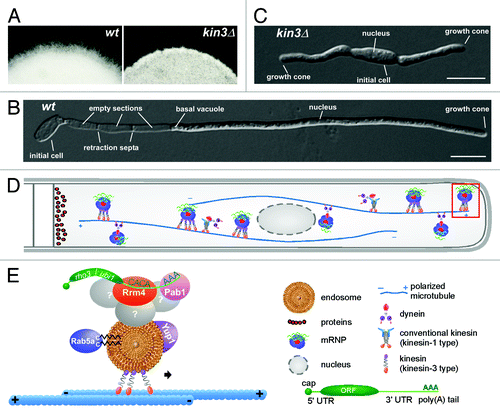
Figure 1. Microtubule-dependent shuttling of Rab5a-positive endosomes. (A) Yeast and (B) filamentous form of strain AB33Rab5aG expressing an active bW2/bE1 variant under the control of a nitrogen-source regulated promoter and the green fluorescent protein (Gfp) fused to the N-terminus of Rab5a (filamentous growth was induced by changing the nitrogen source of the medium; size bars, 10 µm). Rab5aG-positive endosomes (arrowheads in the inverted image detecting Gfp fluorescence) shuttle bi-directionally along microtubules (kymograph in the lower panel). In the kymograph time is plotted vs. distance. Thus, motion of Rab5aG is visible as defined tracks (note the highly processive movement and the reversal of shuttling at the poles). (C) Model depicting the motor-dependent mechanism (three motor system) of endosome transport (endosomes carry the small G protein Rab5a and the SNARE Yup1; symbols are explained in the inlay).
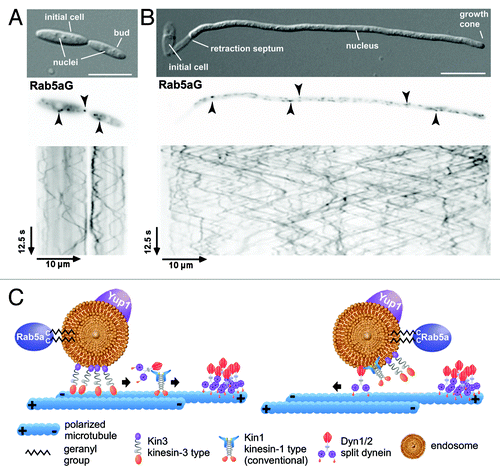
Figure 2. Rab5a-positive endosomes are essential for cytokinesis. (A) Colony and (B) yeast cell morphology of strain AB33kin3Δ in comparison to the progenitor strain (size bar, 10 µm). (C, D) Model depicting the function of Rab5a-positive endosomes in the delivery of Don1 to the site of septation, a process essential for formation of the secondary septum. The red rectangle shown in the middle panel of C is enlarged in D.
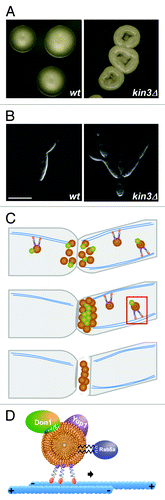
Figure 3. Rab5a-positive endosomes are important for filamentous growth. (A) Colony and (B, C) cell morphology of kin3Δ filaments. The edges of colonies growing under filament-inducing conditions are shown in (A). Monokaryotic filament carrying wild type allele of kin3 (B) is compared with the bipolarly growing filament of kin3Δ (C) strain grown under filament-inducing conditions (size bars, 10 µm). (D, E) Model depicting the function of Rab5a-positive endosomes during transport of mRNPs. Red rectangle shown at the growth cone is enlarged in E (symbols are explained in the inlay).