Figures & data
Figure 1.M. phaseolina infected seedlings after two days of infection. (A) C. trilocularis seedlings, sclerotia attached to the root (B) Both intra and inter cellular Hyphal growth in root and subsequently growth of sclerotia in C. olitorius seedlings (C) New sclerotia (dark spots) observed in infected C. olitorius seedlings (D) Conidia formation and destruction of the host tissue (in C. olitorius)
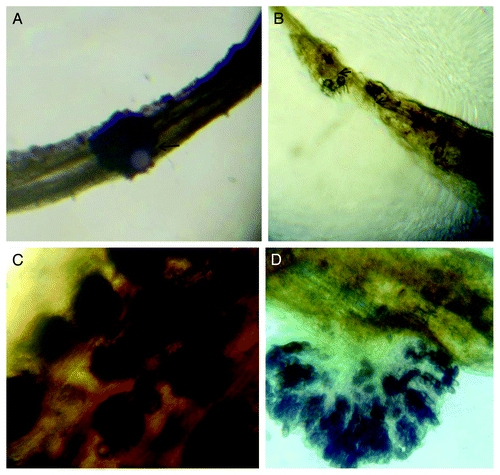
Figure 2. Schematic diagram of CtXTH1 and CoXTH1 gene structure, the white boxes represent exons, lines introns and the shaded boxes are the putative signal peptide sequences.
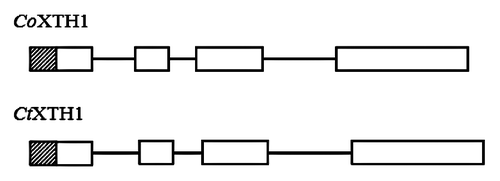
Figure 3. Phylogenetic relationship of CoXTH1 and CtXTH1 with Arabidopsis XTH genes. The tree was generated by MEGA 4.1 using neighbor-joining method and p-distance correction. The Arabidopsis genes from Group 1, 2 and 3 are shown in blue, green and red, respectively.
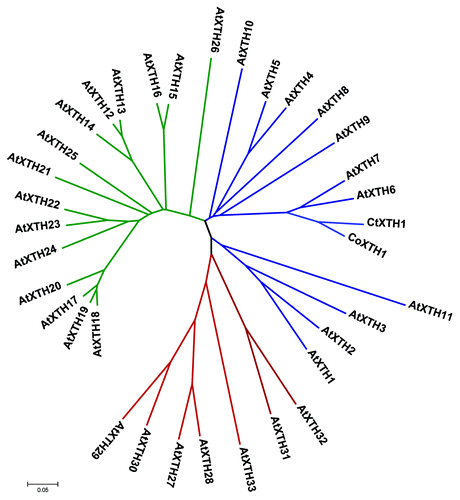
Figure 4. Alignment of the amino acid sequences of seven Arabidopsis group I XTH genes and CtXTH1 and CoXTH1, constructed using ClustalW program and drawn with the Boxshade program. The DELDFELFG and DWATRGG motifs are underlined.
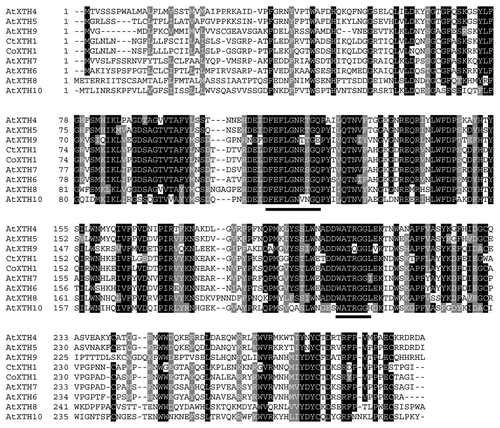
Figure 5. 3D model of CtXTH1 constructed using CPHmodels-3.0 server and represented using Swiss-Pdb Viewer showing typical β-jellyroll-type structure.
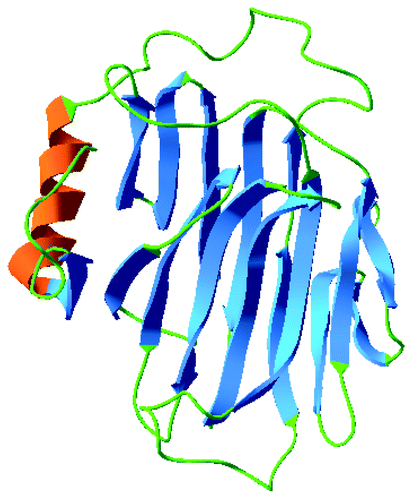
Figure 6. Southern hybridization analysis of genomic DNA from C. olitorius (Lane 1E and 2B) and C. trilocularis (Lane 3E and 4B) digested with EcoR I (E) and BamH I (B).
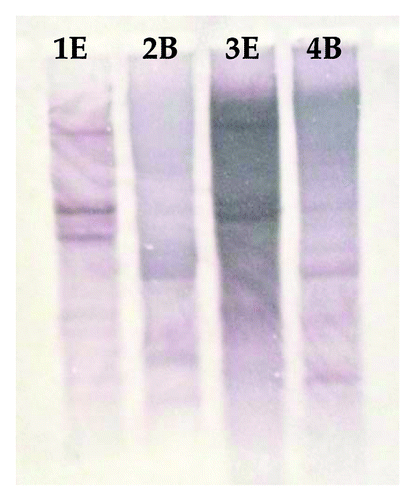
Figure 7. (A) Semi-quantitative Reverse Transcription-PCR expression analysis of three C. olitorious samples (S0, S15 and S24) and three C. trilocularis samples (T0, T15 and T24) using XTHG'F and XTH 3′R primers. (B) The housekeeping gene β-actin was used to verify the quantitation.
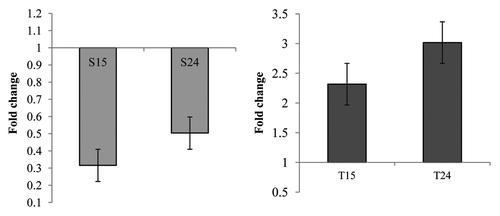
Figure 8. Real-time PCR analysis of (A) CoXTH1 and (B) CtXTH1 genes to see the change in expression level by M. phaseolina. Samples were infected and collected after 15 (S15 and T15) and 24 (S24 and T24) hours of incubation. The mean fold changes of expression (of two duplicate assays) using RNA samples from infected seedlings shows the downregulation of CoXTH1 and upregulation of CtXTH1 followed by fungal infection.