Figures & data
Figure 1 Phylogenetic analysis of Norbin sequences. Full length Norbin amino acid sequences (FASTA format) were aligned using the program MUSCLE. The phylogenetic analysis was performed on this alignment using PhyML and the tree was drawn using TreeDyn. The GeneID or accession numbers corresponding to each of the sequences used are indicated between parentheses. The mammalian species were grouped (red box) and were analyzed independently to increase the resolution. The class to which the species presented belong is indicated on the right in italics.
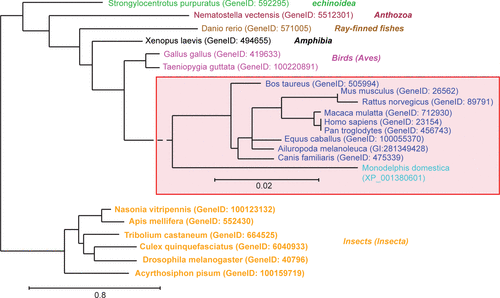
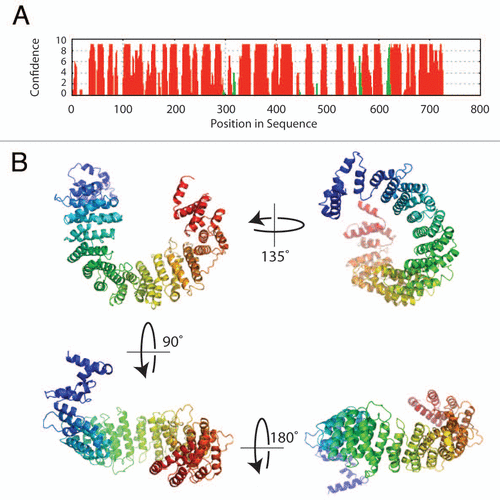
Figure 2 Sequence analysis and structure prediction of Norbin. (A) Secondary structure prediction of Norbin. The full length amino acid sequence of Norbin was analyzed using PsiPReD (v2.6). (B) Norbin helical fold architecture. The helical HEAT repeats detected in the Norbin sequence by fold recognition methods were used to build a plausible three-dimensional structure of human Norbin using comparative modeling techniques.Citation16 The seventeen helical repeats shown in (B) (colored in a blue-to-red N-terminal to C-terminal gradient, respectively) form a curved structure with a well defined inner (or concave) surface that forms the likely area of protein interaction.