Figures & data
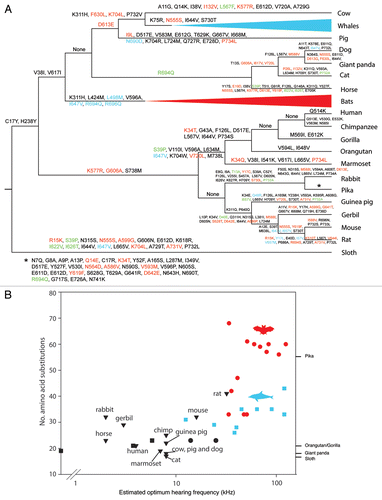
Figure 1 (A) Prestin amino acid substitutions mapped onto the evolutionary path for each taxon. Red amino acid substitutions are convergent with one or more lineages of echolocating bat, blue substitutions are convergent with one or more lineages of echolocating whale and green are convergent with both of these groups. Changes along the pika branch are shown by an asterisk. Names and sequence details of echolocating taxa, cow, pig, horse, dog, cat, human, rabbit, gerbil, mouse and rat are listed in ref. Citation6. Other mammal sequences were obtained from GenBank (giant panda XM_002928662, opossum XM_001371300 and platypus XM_001507913, with the latter two used as outgroups) or from Ensembl using BLAT. (B) Number of prestin amino acid substitutions versus estimated frequency of peak hearing sensitivity for mammals for which audiogram data were available (listed in ref. Citation6 and refs. Citation12—Citation24). Note that the elephant's hearing data is based on the Indian elephant, whereas the gene sequence is from the African elephant. Red and black circles represent echolocating and non-echolocating bats, respectively, and blue and black squares are echolocating and non echolocating baleen whales, respectively. Non-echolocating mammals are shown as black triangles. Species for which auditory data were not available are listed on the right-hand axis..