Figures & data
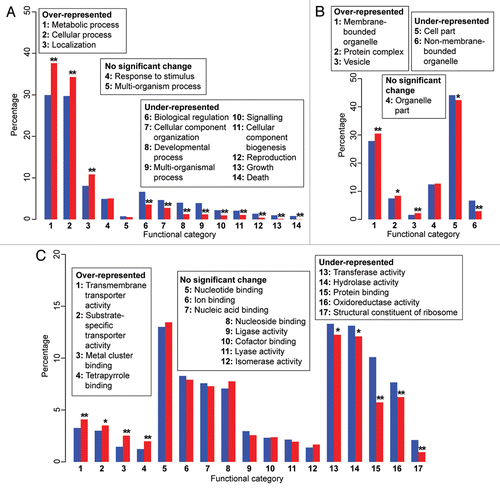
Figure 1 Annotated functional categories of proteins encoded by Porphyridium cruentum. The charts are organized based on three principal categories in the Gene Ontology database: (A) biological process, (B) cellular component and (C) molecular function. The blue bars show the proportional representation of the functional categories (in percentage) across the overall dataset (8,082 annotated genes), whereas the red bars show the proportional representation of the same functional categories (in percentage) among the 1,808 genes implicated in E/HGT. The categories are numbered independently for each panel, as shown in the legends. Significance of over- or under-representation is represented by single (p ≤ 0.05) and double asterisks (p ≤ 0.01), as inferred based on the approach of Chan et al.Citation17