Figures & data
Figure 1 Structures of chromodomains from the HP1 protein (Drosophila melanogaster; pdb:1q3l)Citation4 and PpatensLTR1 (Gypsy LTR retrotransposon from moss Physcomitrella patens) (A); and multiple alignment of ‘classical’ chromodomains and ‘shadow’ chromo-like motifs from functional proteins as well as group I and group II Gypsy LTR retrotransposon chromodomains (B). The PpatensLTR1 chromodomain structure was predicted by (PS)2.Citation5 The residues that constitute the aromatic pocket are indicated by arrows at the top. The secondary structural elements are shown at the bottom: arrows indicate β-strands; cylinder indicates α-helix. The GenBank accession numbers for the sequences are as follows: DmHP1—M57574; DmPc—P26017; MoMOD1 and MoMOD1scdv—P23197; hSUV39H1—S47004; HsHP1scd—AAB26994; DmMi2-2—AF119716; MAGGY—L35053; MGRL3—AF314096; Pyret—AB062507; Cft1—AF051915; PpatensLTR1—XM_001752430; sr35—AC068924; rn8—AK068625; RIRE3—AC119148; Tekay—AF050455; RetroSor2—AF061282; Tma—AF147263.
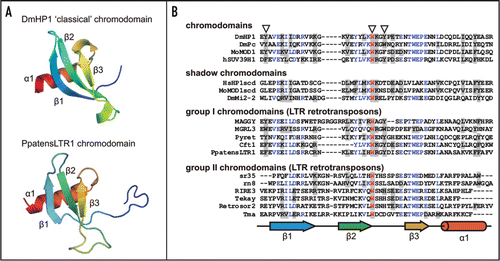
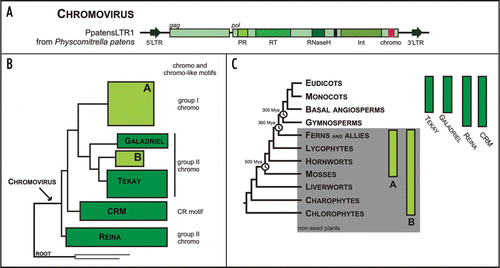
Figure 2 Chromoviruses in plants: structural organization of PpatensLTR1 from Physcomitrella patens (A); a schematic evolutionary tree of plants chromoviruses reconstructed from a multiple alignment of reverse transcriptase DNA fragment (B); and distribution of various chromovirus clades (C). Abbreviations: 5′ and 3′ LTRs—5′ and 3′ long terminal repeats; PR—aspartyl protease, RT—reverse transcriptase, RNaseH—ribonuclease H, Int—integrase, chromo—chromodomains. The plant phylogenetic tree is after Bowman et al.Citation40 with minor modifications. The divergence time estimations are shown after Hedges.Citation41 For a more detailed evolutionary tree of plant chromoviruses, see Novikova et al.Citation42