Figures & data
Figure 1. Growth properties of trs23 mutations. (A) MSY62 was transformed with a plasmid (pRS315) containing TRS23 that was subjected to either random mutagenesis yielding nonsense mutations after the 17th, 52nd and 120th amino acids (trs23Δ202C, trs23Δ167C and trs23Δ99C, respectively); or site-directed mutagenesis to generate a nonsense mutation after amino acid 110 (trs23Δ109C) or deletion of either the first 15 amino acids (trs23Δ15N) or the SMS domain (trs23ΔSMS). Cells were plated on either SD-leucine or on 5-FOA to counterselect against wild-type TRS23 in the URA3-containing plasmid pRS316 and incubated at 30°C. (B) Serial 10-fold dilutions of wild-type or yeast strains with the mutations indicated were plated on YPD and incubated at either permissive temperature (30°C) or the restrictive temperatures of 16°C or 38°C. (C) A schematic showing all of the TRS23 mutations used in this study with their viability and growth phenotypes indicated (cs, cold sensitive; ts, heat sensitive). (D) Human TRAPPC4 (accession number NP_057230.1) and S. cerevisiae Trs23p (accession number NP_101532.1) were aligned using Clustal W and the gaps were inserted manually in accordance with Kim et al., 2006. Identities are shaded in black, the PDZ-like domain of human TRAPPC4 is underlined and the SMS domain of Trs23p is shaded in gray.
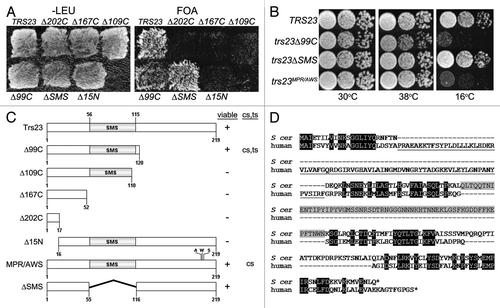
Figure 2. Assembly and function of recombinant TRAPP containing the trs23Δ99C and trs23ΔSMS proteins. (A) Bacterial cells expressing wild-type TRAPP I proteins (with His-tagged Bet3p and Trs33p) and Trs23p (lanes 1–3) or trs23Δ99C (lanes 4–6) were probed for Trs23 using anti-Trs23p antiserum either in the total cell lysate (TCL; lanes 1 and 4), the soluble fraction (lanes 2 and 5) or in the eluate following a Ni2+-NTA agarose purification (lanes 3 and 6). (B) Bacterial cells expressing wild-type TRAPP I proteins (where Bet3p is His-tagged) and Trs23p, trs23Δ99C or trs23ΔSMS as indicated, were first purified with Ni2+-NTA agarose and then the eluates were fractionated by size exclusion chromatography on a Superdex 200 column. Fractions from the column were then analyzed by SDS-PAGE and the gel was silver stained. The positions of the TRAPP proteins are indicated to the side of each panel and the position on the column of fully-assembled recombinant TRAPP I is indicated by an asterisk (*). (C) Lysates from the bacterial cells in (B) were assayed for Ypt1p GEF activity as described in Experimental Procedures [wild-type (■); trs23Δ99C (●); trs23ΔSMS (○)]. The intrinsic ability of Ypt1p to release nucleotide is also shown (Δ). Assays represent three replicates and error bars represent ± SEM.
![Figure 2. Assembly and function of recombinant TRAPP containing the trs23Δ99C and trs23ΔSMS proteins. (A) Bacterial cells expressing wild-type TRAPP I proteins (with His-tagged Bet3p and Trs33p) and Trs23p (lanes 1–3) or trs23Δ99C (lanes 4–6) were probed for Trs23 using anti-Trs23p antiserum either in the total cell lysate (TCL; lanes 1 and 4), the soluble fraction (lanes 2 and 5) or in the eluate following a Ni2+-NTA agarose purification (lanes 3 and 6). (B) Bacterial cells expressing wild-type TRAPP I proteins (where Bet3p is His-tagged) and Trs23p, trs23Δ99C or trs23ΔSMS as indicated, were first purified with Ni2+-NTA agarose and then the eluates were fractionated by size exclusion chromatography on a Superdex 200 column. Fractions from the column were then analyzed by SDS-PAGE and the gel was silver stained. The positions of the TRAPP proteins are indicated to the side of each panel and the position on the column of fully-assembled recombinant TRAPP I is indicated by an asterisk (*). (C) Lysates from the bacterial cells in (B) were assayed for Ypt1p GEF activity as described in Experimental Procedures [wild-type (■); trs23Δ99C (●); trs23ΔSMS (○)]. The intrinsic ability of Ypt1p to release nucleotide is also shown (Δ). Assays represent three replicates and error bars represent ± SEM.](/cms/asset/3ba20e50-1cfd-4d71-a79c-f24d759e11a1/kcll_a_10919414_f0002.gif)
Figure 3. The SMS domain of Trs23p contributes to the strength of Trs23p-TRAPP subunit interactions. (A) Wild-type Trs23p or the mutants trs23Δ99C and trs23ΔSMS were expressed in yeast as fusion proteins with the DNA binding domain of Gal4p and mated to yeast strains expressing other TRAPP subunits fused to the transcriptional activation domain of Gal4p. Serial 10-fold dilutions of diploids were plated on SD-leucine/tryptophan, SD-leucine/tryptophan/histidine, SD-leucine/tryptophan/adenine and SD-leucine/tryptophan/histidine containing 5mM 3AT and all plates were incubated at 30°C. A two-dimensional schematic of the TRAPP II complex is shown to the right of the panel. Shaded in gray is the TRAPP I core of the complex whose crystal structures and organization are known. The position and interactions of the remaining subunits (stippled) are not known but are based on previously published studies on the organization of yeast and mammalian TRAPP complexes.Citation25,Citation27,Citation36 (B) Lysates were produced from yeast cells expressing Trs23p, or the mutant proteins trs23ΔSMS or trs23Δ99C from the plasmid pGBKT7. Equal amounts of protein were loaded in each lane and probed with anti-myc IgG since the fusion proteins contain a myc epitope tag. Identical results were obtained when the blot was probed with anti-Trs23p IgG (not shown).
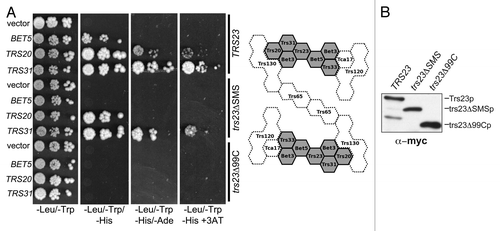
Figure 4. GEF activity is unaffected in S. cerevisiae containing trs23ΔSMS. (A) Lysates from yeast cells expressing wild-type Trs23p (■) or the mutant proteins trs23Δ99C (●), trs23ΔSMS (○) or trs23MPR/AWS (X) were prepared and assayed for Ypt1p-directed GEF activity as described in Experimental Procedures. The intrinsic ability of Ypt1p to release nucleotide is also shown (▾). (B) Lysate from wild-type (■) or trs23ΔSMS (○) was fractionated by size exclusion chromatography on a Superose 6 column in 150mM salt. The fractions enriched in TRAPP II/III (fractions 16–17) and TRAPP I (fractions 29–31) were pooled, concentrated and assayed for Ypt1p GEF activity compared with the intrinsic ability of Ypt1p to release nucleotide. The intrinsic ability of Ypt1p to release nucleotide is also shown (▾). Assays in (A) and (B) represent three replicates and error bars represent ± SEM.
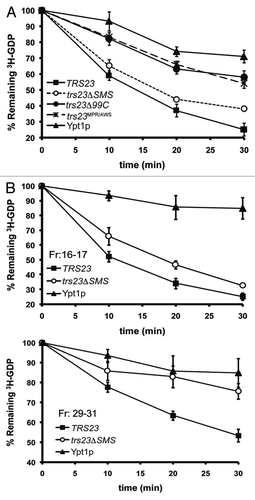
Figure 5. The levels of TRAPP subunits are largely unaffected in trs23ΔSMS. Lysates from wild-type (lane 1), trs23MPR/AWS (lane 2), trs23ΔSMS (lane 3) and trs23Δ99C (lane 4) were probed with antibodies recognizing the TRAPP subunits indicated or anti-HA to detect endogenously-tagged Trs130p. Tubulin served as a loading control.
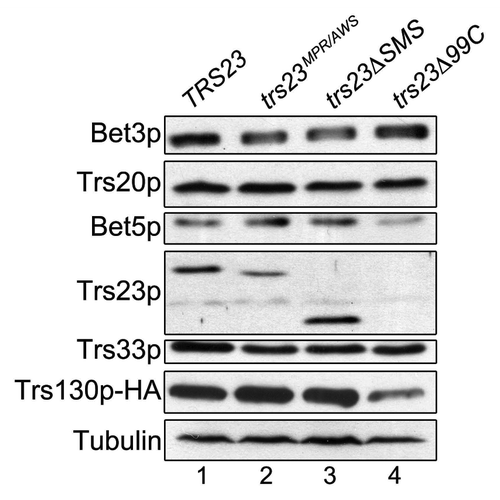
Figure 6. A single TRAPP peak is detected in trs23ΔSMS at physiological salt concentrations. (A) Lysates from wild-type and trs23ΔSMS were fractionated on a Superose 6 size exclusion column in buffer containing either 150 mM NaCl or 300 mM NaCl as indicated. Fractions of 0.5 ml were collected and probed with antibodies that recognize Bet3p, Trs33p, Trs23p or HA (to detect endogenously-tagged Trs130p or Trs85p). I, II and III above the wild-type and trs23ΔSMS blots indicate the location of TRAPP I, II and III, respectively, under conditions where they separate from each other. Molecular size standards are also indicated above the blots. Note that a cross-reactive band appears just above the Trs33p band in fractions 32–34 with the anti-Trs33p antibody and the dash to the right of the Trs33 panels indicates the position of Trs33p. For each subunit shown, samples ranging from fractions 14–38 were fractionated on two separate polyacrylamide gels and processed for western analysis simultaneously. Exposures for each half of the two gels were identical. (B) The signals for Trs33p (top panel) and Trs23p (bottom panel) from wild-type (green) and trs23ΔSMS (blue) at 150 mM NaCl shown in (A) were quantitated using Image J and plotted as a percentage of the total signal. Note the absence of the TRAPP I peak in trs23ΔSMS. (C) Lysates were prepared for fractionation as in (A) at either 150 mM or 300 mM NaCl. Samples before the centrifugation (T), the pellet fraction following centrifugation (P) and the supernatant (S) that was loaded onto the Superose 6 column were probed for Trs23p in both wild-type and trs23ΔSMS. (D) Lysate from wild-type cells containing Trs85p-TAP and Trs130p-myc were fractionated on a Superose 6 column and fractions 16–17 were collected and split in two. One sample was treated with DMSO while the other sample was treated with the crosslinking reagent DSP for 3 h on ice. Each sample was then split in two and either treated with (+) or without (-) anti-TAP IgG. The immune complexes were collected on protein A-sepharose beads, fractionated by SDS-PAGE and probed with anti-myc IgG. Samples representing 10% of the input are shown.
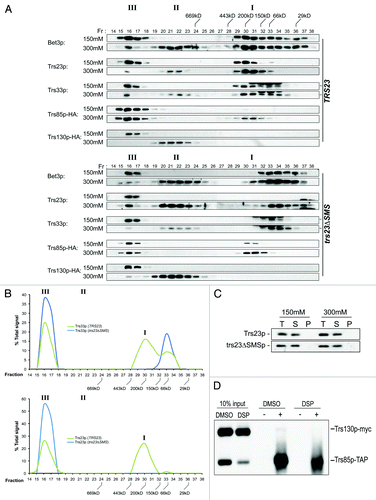
Figure 8. TRAPP I can form from TRAPP II/III. (A) Lysates of wild-type yeast were prepared in buffer containing 50 mM, 150 mM or 300 mM NaCl and fractionated by size exclusion chromatography on a Superose 6 column in buffer containing identical salt. The fractions were analyzed by western blotting for the presence of Trs23p. (B) The TRAPP II/III-containing fraction from wild-type lysate that was fractionated in the presence of 50 mM NaCl was incubated with 2 mM DSP or DMSO, quenched with Tris pH 7.5 and re-fractionated on a Superose 6 column in buffer containing 300 mM NaCl. Fractions were probed for the presence of Trs33p. Note that a cross-reactive band appears just above the Trs33p band in fractions 32–34 with the anti-Trs33p antibody in the DMSO-treated sample and the dash to the right of the Trs33 panel indicates the position of Trs33p. I, II and III above the wild-type and trs23ΔSMS blots indicate the location of TRAPP I, II and III, respectively, under conditions where they separate from each other.
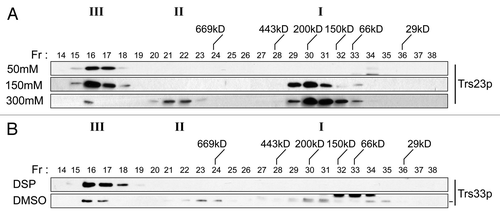
Figure 7. Membrane trafficking is unaffected in the trs23ΔSMS mutant. (A) Wild-type, trs23ΔSMS and sec18 yeast were pulse-labeled with 35S-methionine/cysteine and chased for the times indicated below the panels. Carboxypeptidase Y (CPY) was immunoprecipitated from lysates at each time point, fractionated by SDS-PAGE and visualized by autoradiography. The position of the ER (p1) and vacuolar (m) forms of CPY are indicated. (B) Wild-type, trs23ΔSMS and trs85Δ were transformed with a plasmid expressing GFP-Snc1p. The cells were fixed and viewed using an epifluorescence microscope. (C) Wild-type, trs23ΔSMS and trs23Δ99C were transformed with a plasmid expressing Ypt31p-GFP. The cells were fixed and viewed using an epifluorescence microscope. For quantitation in (B) and (C), a minimum of 100 cells from three replicates were counted. The error bars represent ± SEM and representative cells used for quantitation are shown as insets. (D) Wild-type, trs23ΔSMS and trs85Δ were transformed with a plasmid expressing Ape1p-GFP, grown in minimal medium without uracil, converted to spheroplasts and lysed. Samples were fractionated by SDS-PAGE and probed with anti-GFP. The percentage of Ape1p-GFP processing, calculated using Image J, is indicated below each lane. (E) Reconstitution of ER-to-Golgi traffic was performed as described in Experimental Procedures. Reactions were performed with no additions either on ice (ice) or at 29°C (NA), or fully reconstituted at 29°C (Recon). The results are expressed as percentage of concanavalin A-precipitable pro-α-factor that has received α-1,6-mannose Golgi modifications. The results are the average of duplicates and the range of transport over three independent experiments was 13.6–15.4% (wild-type) and 12.6–14% (trs23ΔSMS). (F) Lysates were prepared from wild-type and trs23ΔSMS in buffer B88 that was used for the in vitro transport assay in (E) and fractioned on a Superose 6 column in B88. Fractions were probed with anti-Trs33p. I, II and III above the blots indicate the location of TRAPP I, II and III, respectively, under conditions where they separate from each other.
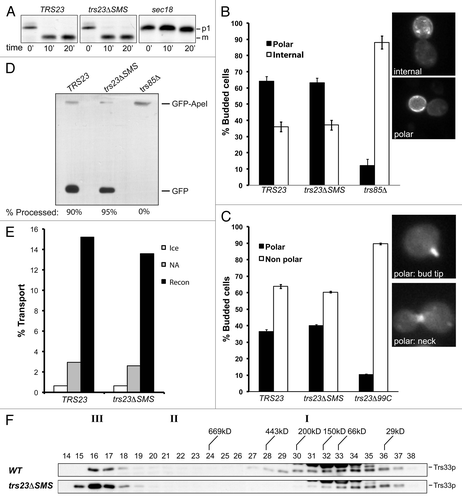
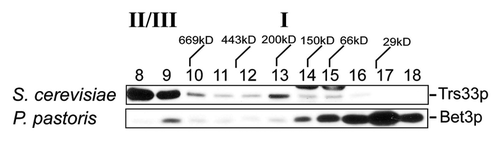
Figure 9.P. pastoris does not contain a TRAPP I peak. Lysates in 150 mM NaCl from P. pastoris and S. cerevisiae were fractionated by size exclusion chromatography on a Superdex 200 column. The fractions were probed for Trs33p (S. cerevisiae) or P. pastoris Bet3p using anti-TRAPPC3 IgG. The positions of TRAPP II/III and I are indicated above the top panel.