Figures & data
Figure 1. Protein and mRNA expression of Wnt5a in non-metastatic human colon cancer cell line SW480 and highly metastatic cell line SW620. (A) Relative mRNA expression of Wnt5a in SW480 and SW620 presented as the ratio to the housekeeping gene L7a (n = 3). mRNA was analyzed using real-time RT-PCR. The values are presented as the mean ± SEM *p < 0.05 when comparing SW620 to SW480. (B) Wnt5a protein expression in SW480 and SW620 using immunofluorescent staining. Wnt5a protein was analyzed after immunofluorescent staining using an antibody against Wnt5a and an Alexa Fluor 647-labeled secondary antibody (red). The coverslips were also counterstained with Hoechst 33342 fluorescent stain to detect the nucleus (blue). The two pictures were overlaid to show Wnt5a staining on top of the nuclear staining (merged). (C) Quantification of immunofluorescent staining of Wnt5a (stained in red) in SW480 and SW620. Over 20 cells per slide were randomly chosen for quantification. Data was shown as relative intensity per pixel based on AxioVision4.7 software. The values are presented as the mean ± SEM, *p < 0.05 when comparing SW620 to SW480.
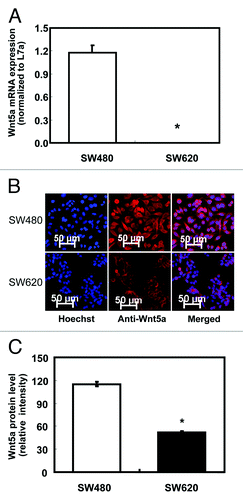
Figure 2. Expression of Wnt5a mRNA after 5-aza-cytidine (Azacitidine), trichostatin A (TSA) and sodium butyrate (NaBt) treatment. (A) Relative Wnt5a mRNA level was tested in SW480 and SW620 treated with Azacitidine (0, 4 and 8 μmol/L) for 48 h. mRNA was analyzed using real-time RT-PCR and presented as the ratio to L7a (n = 3). (B) Wnt5a mRNA level was tested in SW620 treated with TSA (50 and100 nmol/L) or NaBt (2.5 and 5 mmol/L) for 48 h. mRNA was analyzed using real-time PCR and presented as the ratio to L7a (n = 3). The values are presented as the mean ± SEM, *p < 0.05 when compared with the DMSO-treated SW620 control group.
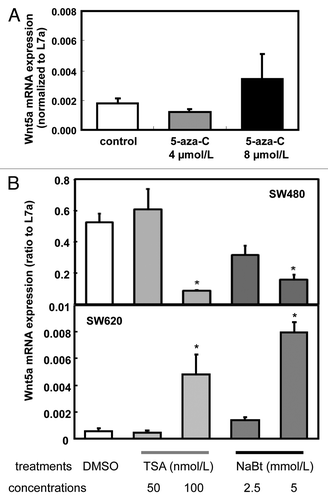
Figure 3. ChIP analysis of chromatin modifications in SW480 and SW620. (A) Histone modifications at the promoter region (-96/-22, upper panel) and coding region (+6424/+6495, lower panel) of human Wnt5a in SW480 and SW620 (n = 3). Data are normalized to the input DNA. The values are presented as the mean ± SEM *p < 0.05 indicates statistical significance compared with SW480. (B) Histone modifications at the promoter region (-96/-22, upper panel) and coding region (+6424/+6495, lower panel) of human Wnt5a in SW620 after NaBt treatment (5 mmol/L) for 48 h (n = 3). Data are normalized to the input DNA. The values are presented as the mean ± SEM *p < 0.05 when comparing to the non-treated SW620 control group.
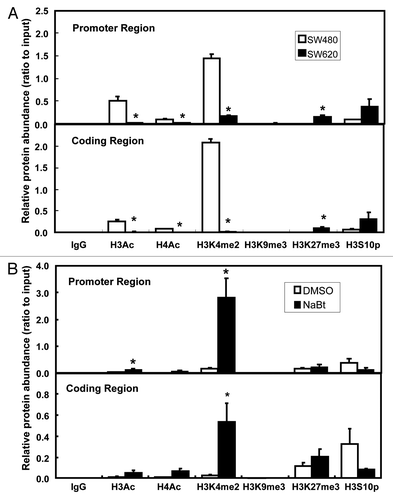
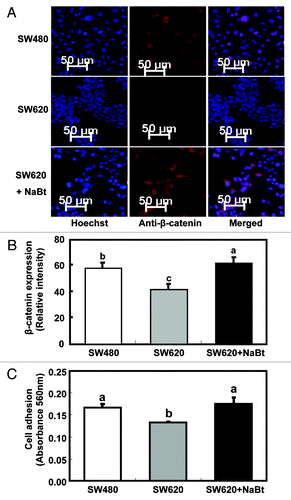
Figure 4. Influence of NaBt treatment on the WNT signaling pathway and cell adhesion ability of SW620. (A) Expression of β-catenin protein in SW480, SW620 and SW620 treated with NaBt (5 mmol/L) for 48 h using immunofluorescence. β-catenin protein was analyzed by immunofluorescent staining using an antibody against β-catenin and an Alexa Fluor 647-labeled secondary antibody (red). The coverslips were also counterstained with Hoechst 33342 fluorescent stain to detect the nucleus (blue). The two pictures were overlaid to show β-catenin staining on top of the nuclear staining (merged). (B) Quantification of immunofluorescent staining of β-catenin protein (stained in red) in SW480, SW620 and SW620 treated with NaBt. Over 20 cells per slide were randomly chosen for quantification. Data was shown as relative intensity per pixel based on AxioVision4.7 software. The values are presented as the mean ± SEM. Values with different letters are statistically different (p < 0.05). (C) Cell adhesion ability of SW480, SW620 and SW620 treated with NaBt (5 mmol/L) for 48 h (n = 3). Cells grown on fibronectin-coated plates were fixed and stained with crystal violet. The numbers of adherent cells were estimated by absorbance reading at 562 nm. The values are presented as the mean ± SEM. Values with different letters are statistically different (p < 0.05).