Figures & data
Figure 1. Schematic of the Kcnq1 imprinted domain on mouse chromosome 7. Arrows indicate direction of transcription. Arrows above the genes represent maternal transcription, below the line paternal transcription and genes with two arrows have biallelic expression. Shown in the light gray box are the genes that are imprinted in both the embryo and placenta. Shown in the dark gray box are the Tnfrsf genes present in the mouse genome but absent in the human. They are transcribed in the same orientation as Cars.
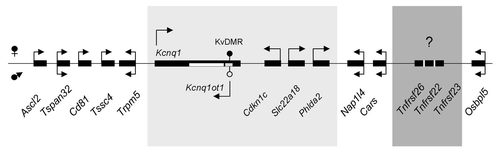
Figure 2. Expression profiles during development for the murine Tnfrsf genes. Top, schematic of the three Tnfrsf genes and their linear organization. Arrows above the line indicate transcriptional direction of Tnfrsf genes, arrow below the line, direction of transcription for AK155734. Cen, centromere; tel, telomere. The Kcnq1 and Kcnq1ot1 genes are telomeric to the Tnfrsf genes. Gray boxes, Tnfrsf exons, white boxes, predicted AK155734 exons. Dark arrows indicate the PCR primers used for the Tnfrsf genes, and white arrows indicate PCR primer sets (A, B, C, D, and Ef/Er) used to determine the presence and splicing of the antisense AK155734 (primer sequences in Table S1). Below, expression levels of each of the indicated Tnfrsf genes relative to Gapdh. ES, mouse embryonic stem cells, 7.5, whole embryos at 7.5 dpc; 10.5 B, 13.5 B, 16.5 B, bodies of embryos at 10.5, 13.5 and 16.5 dpc; nnH, neonatal hearts. Error bars represent standard deviations.
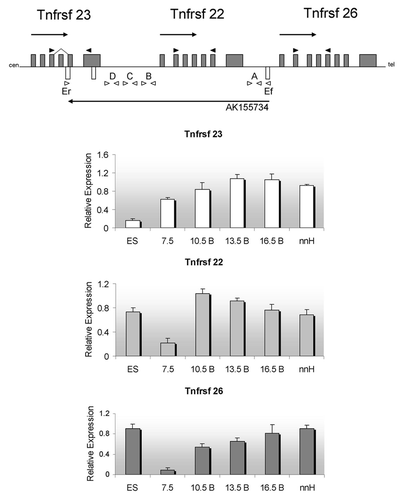
Figure 3. Parent-of-origin expression pattern of murine Tnfrsf genes and the antisense AK155734. (A) RNAs from F1 hybrid E13.5 were subjected to RT-PCR and restriction digests. Allele-specific bands were quantified and the ratio of paternal to maternal transcript was determined. (B) Neonatal heart RNAs from F1 hybrid mice were subjected to RT-PCR and restriction digests, as in A, and allelic ratios were determined. For both (A and B), the results of progeny of C57BL/6J x B6(CAST7) crosses are shown. The reciprocal crosses yielded similar results.
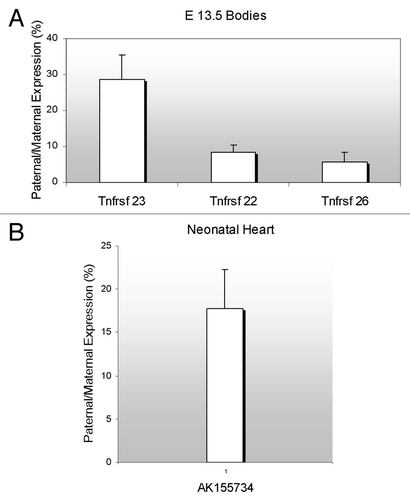
Figure 4. Detection of an antisense non-coding RNA at the Tnfrsf locus. Top, UCSC browser screen of the three Tnfrsf genes. Dark gray block arrow signals the reported antisense gene, AK155734. Below, expression relative to Gapdh of the AK155734 RNA, using primer set A (from ) for embryonic stem cells (ES), whole embryos at E7.5, bodies from embryos (B) at E10.5, 13.5 and 16.5 and neonatal heart (nnH).
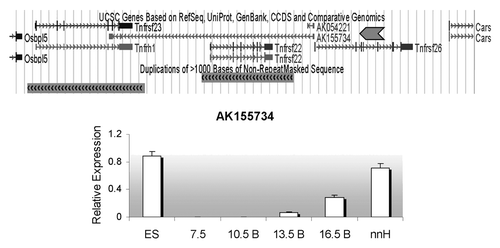
Figure 5. Distribution of Tnfrsf homologous sequences in Kcnq1 orthologous regions. The topology is drawn according to www.tolweb.org/tree and Murphy et al.Citation15 The total numbers of Tnfrsf sequences observed in each taxon are indicated, even in those in which no Tnfrsf homologs are detected, in order to reflect both gene gains and losses within the Kcnq1 region. The figure also illustrates how many of them correspond to orthologs of Tnfrsf22 or Tnfrsf23 and of Tnfrsf26 only for placental mammals, according to the phylograms depicted in and . Only mouse, human and frog Kcnq1 orthologous regions are devoid of sequencing gaps that result from incomplete sequence assembly. #In pig, three identical sequences were found; they could be the result of assembly errors or very recent duplications and, therefore, only one was analyzed in our phylogenetic trees. +In guinea pig, multiple sequences with significant similarity to mouse Tnfrsfs were found; we manually annotated a minimum of nine. *In humans, we only detected one sequence that is much shorter than any of the ones found in other species (Sup. Material). †In wallaby, we could not confirm the location of the Tnfrsf homologous sequences within the Kcnq1 orthologous region (see Materials and Methods).
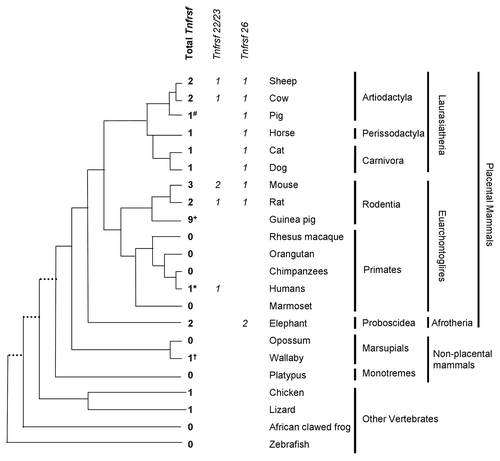
Figure 6. Phylogeny of Tnfrsf homologous sequences within Kcnq1 orthologous regions. The evolutionary history was inferred using the Neighbor-Joining method. Nucleotide sequences that aligned unambiguously to mouse Tnfrsf22, Tnfrsf23 or Tnfrsf26 were analyzed (see Materials and Methods); nine guinea pig sequences were removed from this analysis for the sake of clarity, although similar results were obtained in trees that included them (data not shown). This is a rooted phylogram obtained using MEGA 5.05.Citation13 The evolutionary distances were computed using the Maximum Composite Likelihood method and are in the units of the number of base substitutions per site. Numbers at nodes represent bootstrap support values based on 5000 pseudoreplicates. Values below 50% were removed and * indicates 70% or above bootstrap support.Citation16
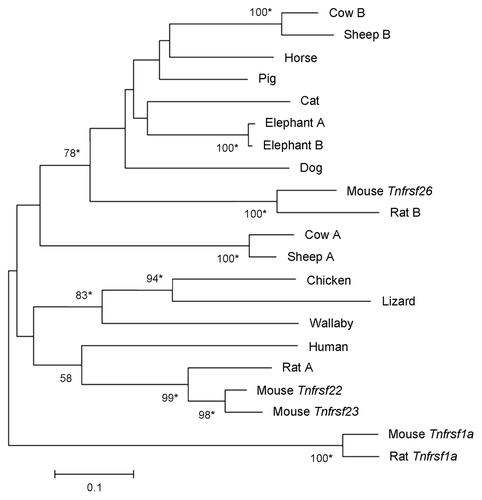
Figure 7. Phylogeny of Tnfrsf homologous sequences within Kcnq1 orthologous regions of placental mammals. The phylogeny was based on the Neighbor-Joining analysis of nucleotide sequences. Analysis was restricted to placental mammalian sequences that aligned unambiguously to mouse Tnfrsf22, Tnfrsf23 or Tnfrsf26, with the exception of human (due to the small size of the orthologous sequences) and guinea pig (see Materials and Methods). This rooted phylogram was obtained using the Maximum Composite Likelihood method implemented in MEGA 5.05.Citation13 The tree is drawn at a scale that represents the number of base substitutions per site. Numbers at nodes represent bootstrap support values based on 5000 pseudoreplicates. Values below 50% were removed and * indicates 70% or above bootstrap support.Citation16
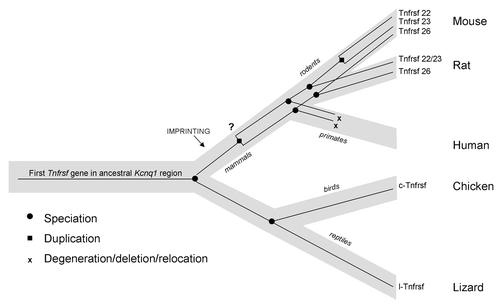
Figure 8. Model of the evolution of mouse and human Tnfrsf genes within the Kcnq1 region. Light gray line represents the species tree; thin black lines within represent the Tnfrsf gene tree. This figure represents only relevant lineages and species in order to summarize the major events in evolution of mouse and human Tnfrsf genes. Tnfrsf sequences were present in the Kcnq1 ancestral region before the first establishment of imprinting in mammals (notice that imprinting acquisition did not occur for all genes at the same time and it remains to be determined when parental bias was established in the Tnfrsf genes). Two duplications (one early or prior to mammalian evolution and one after the mouse and rat lineage split) originated the three Tnfrsf genes present in mouse. It has not been established whether the earlier duplication occurred before or after imprinting emerged in mammals (indicated with a question mark). In primates, Tnfrsf genes degenerated or were deleted or relocated, and in humans only a single short homologous sequence is observed.