Figures & data
Figure 1. Detailed DNA methylation analysis of the ID4 gene 5′ regulatory region. (A) Schematic representation of the ID4 5′ regulatory region including the adjacent CpG island (CGI). The amplicon analyzed by the MassARRAY (MA) assay and by bisulfite sequencing (BS) in AML samples (green) and healthy controls (blue) is depicted as black bar including the position relative to the transcriptional start site (TSS). The regions analyzed by pyrosequencing and methylation-specific PCR are labeled Pyro and MSP (U for reaction with primers specific for unmethylated DNA, M for methylated), respectively. Quantitative MassARRAY results are displayed as heatmap, columns represent single CpG units, rows represent samples. Bright green encodes for low methylation levels, dark blue for high methylation levels. (B) Distribution of average amplicon methylation values by MassARRAY (mC) in AML samples (AML), pooled healthy controls (Ctrl) and separated healthy controls (BM, whole bone marrow; CD34+, G-CSF mobilized CD34 positive hematopoietic progenitor cells from peripheral blood; PB, buffy coat from peripheral blood). Significance was assessed by two-sided non-parametric Wilcoxon Mann-Whitney test. (C) Scatterplot displaying the correlation for mean methylation levels of CpG dinucleotides asessed by both MassARRAY and pyrosequencing. The correlation is calculated as Pearson's R2. (D) Gel electrophoresis of a representative MSP analysis at the ID4 gene 5′ region in AML samples (green) and healthy controls (blue) demonstrating the different MSP-based methylation categories. The location of the amplicon is indicated in . U and M represent the reaction for unmethylated and methylated DNA, respectively. Peripheral blood mononuclear cells (PB), in vitro methylated DNA (IVD) and water (H2O) served as controls. Sample source (BM, PB) and samples IDs are given above the horizontal bars together with the determined methylation category in bold (U, unmethylated; W, weakly methylated; M, methylated). (E) Pie chart showing distribution of MSP-derived DNA methylation categories in healthy controls (n = 18) and AML samples (n = 62).
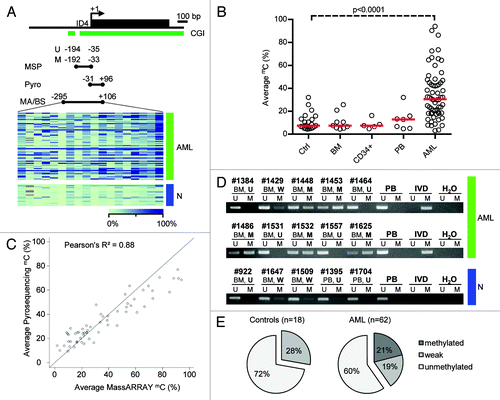
Figure 2. Direct comparison of quantitative MassARRAY and MSP-based methylation data at the ID4 5′ region. (A) Distribution of MassARRAY-derived DNA methylation values (mC) for the MSP categories U (unmethylated), W (weakly methylated) and M (methylated) for single CpG units located inside the MSP primer sequences. The MSP forward primer harbors 3 CpG units (identical with MassARRAY CpGs 7–10), for the MSP reverse primer one CpG unit (CpG 32) can be addressed. (B) Distribution of MassARRAY derived DNA methylation values for the MSP categories U, W and M. The CpG units located in the MSP primer sequence are summarized as mean value. All three MSP groups are tested for difference of location using the non-parametric Kruskal-Wallis test. (C) Determination of a MassARRAY based cut-off to classify samples as methylated generating identical proportions of methylated samples for both methods. The Y-axis describes the percentage of samples defined as being methylated. The black curve displays MassARRAY-derived methylation data, the red line represents MSP-based methylation data leading to a cut-off of 42%. (D) Agreement between both methods (inter-rater reliability) by Cohen’s kappa including confidence interval depending on quantitative MassARRAY data.
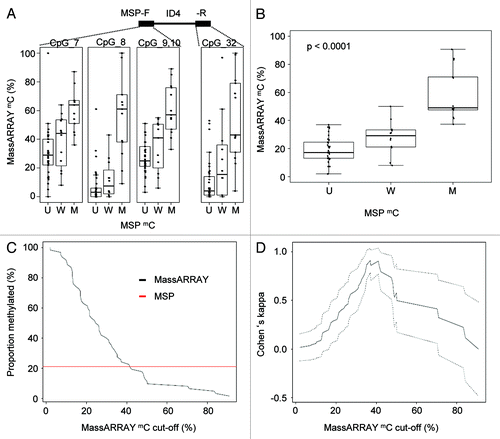
Figure 3. Quantitative DNA methylation analysis of SFRP family members. (A-D) Distribution of MassARRAY-derived DNA methylation values (mC) for the MSP categories U (unmethylated) and M (methylated) for SFRP1 (A), 2 (B), 4 (C) and 5 (D). Distributions of samples categorized as either U or M by MSP were assessed for the average of all CpG units located in MSP primer sequences (indicated as mean F+R). For SFRP1, MSP primers are located in direct vicinity outside of the MassARRAY amplicon, therefore, the mean of the entire MassARRAY amplicon was used for comparison. (E-H) Agreement (inter-rater reliability) between both methods by Cohen’s kappa including confidence interval depending on quantitative MassARRAY data (mC) of all CpG units located in MSP primer sequences (indicated as mean F+R in A-D).
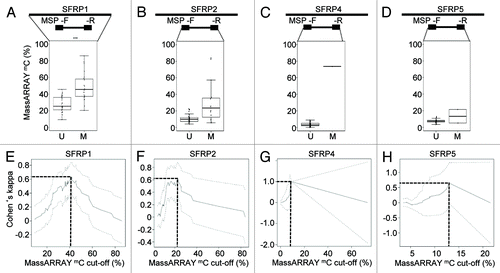
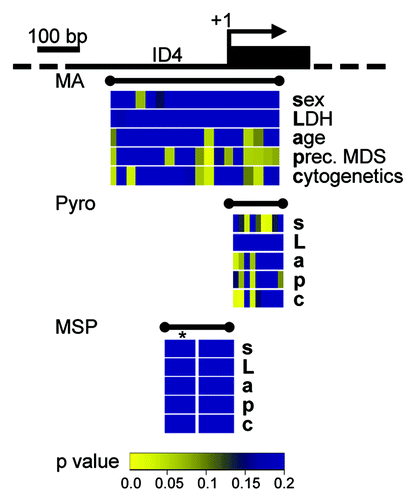
Figure 4. Quantitative DNA methylation data of the ID4 5′ region reveal correlations with AML disease characteristics. Graphical display of the ID4 gene 5′ region and all raw non-parametric p-values for single the factors sex (s), LDH (L), age (a), presence of a preceding MDS (p) and cytogenetic risk group (c) as derived from univariate testing. Correlations were performed on single CpG units of MassARRAY-derived methylation data as well as for single CpG methylation values by pyrosequencing (Pyro). P-values are displayed in spatial relation to the transcriptional start site (+1). For correlations with MSP-derived DNA methylation status, MSP data were treated either as binary variable (U/M) or using a separation in three methylation states (U/W/M, indicated by *). Color-coding displays low raw p values (significant low p values are depicted in yellow colors; higher p values are represented in a green-blue transition). All p values > 0.2 are displayed in blue.