Figures & data
Table 1. Baseline characteristics of breast cancer patient cohorts
Figure 1. Methylation frequencies of cancer specific methylated genes in primary BCs. Bar graph showing methylation frequencies (%) of 33 cancer specific methylated genes in the Hopkins training cohort (n = 30).
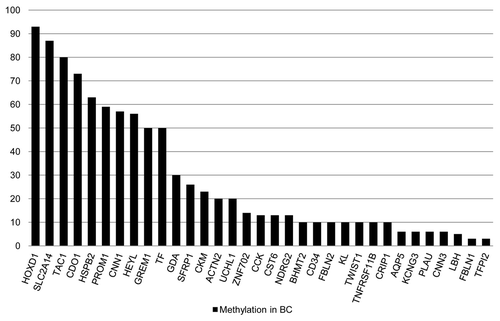
Table 2. Combinatorial beacon marker analysis for detection of breast cancers
Table 3. Statistical association between clinocopathological characteristics and methylation frequencies (%) of genes in Hopkins extended training cohort
Figure 2. Single gene and gene combinations for prognostication of mortality risk in BC patients. (A) Forest plots depicting univariate and multivariate HRs and corresponding 95% CIs for overall mortality risk associated with DNA methylation of single or combinations of genes in Hopkins extended training cohort (n = 97) and (B) Hopkins validation cohort (n = 89). Vertical line at 1 indicates HR = 1. Single gene marker and selected statistically significant gene combinations are shown. (C) Univariate and multivariate Cox proportional hazards regression based survival curves for prognostic gene marker combination TAC1 and CKM in Hopkins extended training cohort and Hopkins validation cohort.
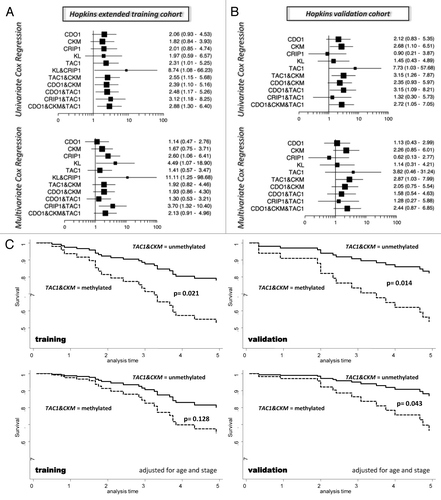
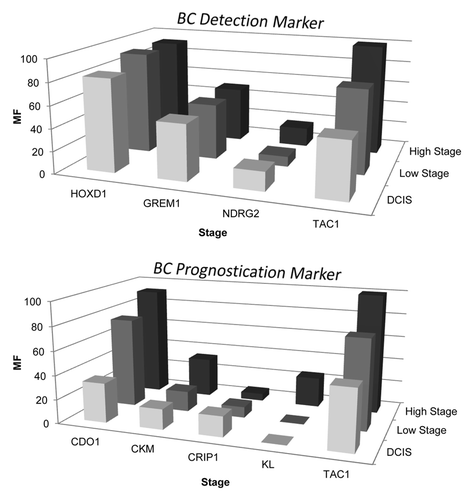
Figure 3. Distinct methylation frequency pattern for BC detection vs. BC prognostication marker. Methylation frequency of single marker genes (GREM1, HOXD1, NDRG2, TAC1, CDO1, CKM, CRIP1 and KL) was plotted against cancer stage as obtained in Hopkins training cohort (n = 30). Two groups, BC detection marker and BC prognostication marker, of genes showing distinct methylation frequency pattern were identified. Upper plot shows BC detection markers (GREM1, HOXD1, TAC1) that are characterized by methylation frequency ≥ 40% in early stage BC’s (DCIS and stage 1). Lower plot shows BC prognostication markers (CDO1, CKM, KL and TAC1) that are characterized by stage increasing methylation frequency. Note NDRG2, an early detection marker, did not reach the 40% methylation frequency cut off in early stage BCs by highly stringent MSP assay and CRIP1, a prognostic marker, did not follow the stage increasing methylation frequency pattern as seen with the other prognostic markers.