Figures & data
Figure 1. Trans-chromosomal methylation (TCM) and trans-chromosomal demethylation (TCdM). In a TCM event in the hybrid, the unmethylated parental allele (Parent B) increases in methylation to resemble the methylated parental allele (parent A) at locus At3g43340. In a TCdM event in the hybrid, the methylated parental allele (parent A) shows a reduction in methylation (mainly associated with CHH methylation) at a region with reduced levels of siRNAs in the hybrid at locus At2g02660. Methylated CG (blue), CHG (red) and CHH (green) are plotted along the x-axis with levels of methylation and siRNAs plotted on the y-axis. TCM and TCdM correlate with regions producing siRNAs (plotted in red above each methylation graph).
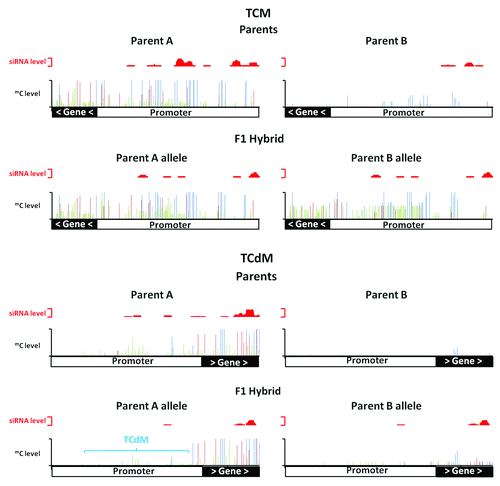
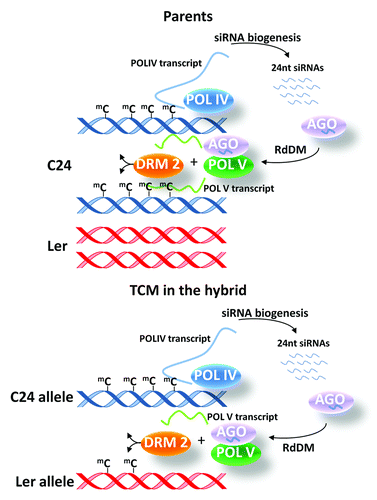
Figure 2. siRNA-mediated trans chromosomal methylation. The RNA directed DNA methylation pathway plays a key role in TCM and TCdM events. RNA Polymerase IV (POL IV) transcribes a region which following double stranded RNA synthesis and cleavage forms 24 nt siRNAs. Twenty-four nt siRNA are loaded into an ARGONAUTE (AGO) and target regions by an RNA transcript produced by RNA Polymerase V (POL V). This complex that contains additional components can then recruit DOMAIN REARRANGED METHYLTRANSFERASE 2 (DRM2), which methylates the DNA. In hybrids, siRNAs derived from the methylated parental allele target both the methylated parental allele for continued methylation (blue helix) and de novo methylation of the unmethylated parental allele (red helix). After the initial TCM event (illustrated here) the previously unmethylated allele will produce siRNAs and transcripts produced by POL V enabling the newly methylated cytosines to be maintained through subsequent generations.