Figures & data
Figure 1. Relationship between DNA methylation and irradiation. Genomic DNA cytosine methylation measured by HPLC-UV of un-irradiated mESC samples and mouse control tissues before (A) and after irradiation (B). Columns represent the average percentage of cytosine bases that were methylated. Error bars represent the standard error of the mean (SEM). Results were combined from multiple independent experiments. The number of replicate measurements for each sample is displayed above the error bar. (C) Dose-response curve of the ESC lines constructed using the long-term clonogenic assay after irradiation with 0–7Gy X-rays. Each data point represents the mean, and error bars represent the SEM, of at least 3 independent experiments. (D) Linear quadratic parameters α and β were calculated by fitting the data displayed in with a linear quadratic equation using SPSS and are shown in columns 3 and 5. Column 2 indicates the Plating Efficiency for each clone and column 4 and 6 the standard errors (SE) for α and β respectively. The survival fraction at 2Gy (SF2) inferred from the data are shown in the last column.
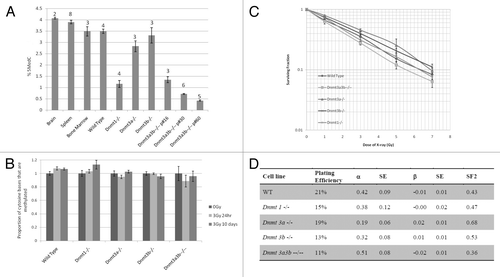
Figure 2. Analysis of single strand, double strand and alkali labile site DNA damage (A) or damage resulting from oxidative stress (B) in wild-type, Dnmt1−/−. Dnmt3a3b–/– mESC lines, 23–25PDs post 3Gy X-irradiation or sham treatment as measured by percentage of tail DNA. Results are expressed as an average of 3 independent experiments, each comprising 200 separate cell measurements. Error bars represent the SEM of the averages of the 3 independent experiments.
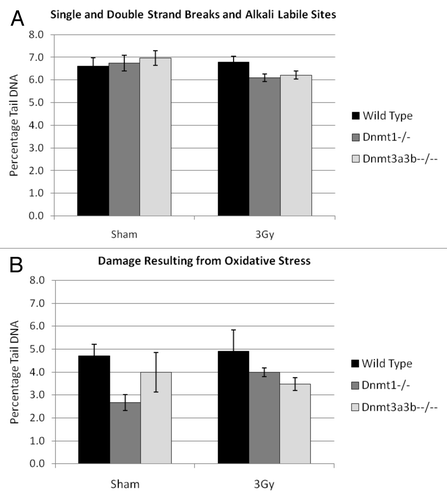
Figure 3. Classifications of structural cytogenetic aberrations as defined by Savage.Citation33 The results are displayed as the number of each class of aberration observed per 100 metaphases. Graph (A) displays the results 10–14 PDs post treatment; Graph (B) displays the results 23–25 PDs post treatment. (C) Mitotic spreads of Giemsa-stained chromosomes from the Dnmt1−/− ESC line 23–25 PDs after 0Gy treatment, showing a chromosome with very long satellite arms. Immediately after exposure to 3Gy X-rays or sham treatment, cells were seeded into the clonogenic assay. Surviving colonies were counted 12–14 d after seeding. (D) Dicentric, tri-centric and tetra-centric chromosome-type aberrations observed in Giemsa-stained metaphase spreads of Dnmt1−/− and Dnmt3a3b–/– mESCs 23–25PDs post 0Gy (sham) or 3Gy treatment.
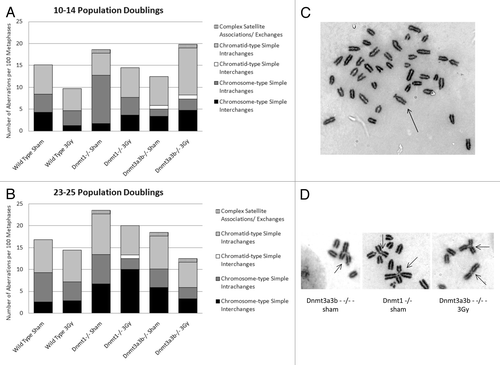
Table 1. Absolute numbers of each cytogenetic aberration type observed at 10-14 PD and 23-25 PD post treatment with 0Gy or 3Gy X-rays
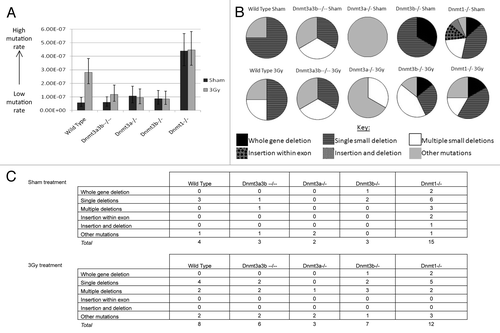
Figure 4. (A) Hprt gene mutation rates 23–25 population doublings after 3Gy X-irradiation or 0Gy (sham) treatment. Columns represent the mutation rate, calculated from 19–25 individual clonal expansions using Luria-Delbrück fluctuation analysis and corrected for plating efficiency. Error bars represent the 95% confidence intervals. Dnmt3a3b–/– mESCs were p#20–26 at the time of selection for mutants. (B) Spectrum of functional Hprt gene mutations observed in mESC lines 23–25 population doublings after 3Gy X-irradiation or 0Gy (sham) treatment. (C) Tables detailing the mutations identified by exonic PCR that were used to construct the pie charts in (B). PCRs were performed using primer pairs designed to amplify each exon (1–9) of the Hprt gene and exon 2 of the unlinked K-ras gene as a control. When a mutation was observed in multiple colonies arising in the same clonal population it was counted only once. ‘Other mutations’ include mutations that could not be detected using the PCR-based method, such as point mutations, frame shift mutations and epigenetic alterations.