Figures & data
Figure 1.Sat2 hypomethylation is not correlated with constitutive Sat2 RNA overexpression in cancer cell lines. (A) The MethyLight assayCitation3 was used to quantify Sat2 methylation on bisulfite-treated DNA isolated from glioblastoma (n = 10), melanoma (n = 14), sarcoma (n = 10) and carcinoma (n = 22) cell lines. Non-tumoral control samples included four hTERT-immortalized cell lines (HFF2hTERT, HEKhTERT, HNEMhTERT, HMEChTERT) and HMSC mesenchymal stem cells. Methylation levels were compared with Sat2 methylation in HFF2hTERT. Bars indicate median values. (B) The Sat2 sequence of chromosome 1 corresponds to GenBank accession number X72623.1. In light gray: Sat2 fragment analyzed after bisulfite treatment of genomic DNA for methylated CpG content determination by sequencing. The 17 CpG sites are numbered. In dark gray: Sat2 fragment amplified by PCR on cDNA for quantitative RT-PCR measurement of Sat2 RNA transcripts. The same region (dark gray) was amplified in ChIP experiments. The HSF1 binding site predicted by tfsearch is underlined. (C) Sat2 methylation level in cancer cell lines derived from sarcoma, melanoma and carcinoma and in HFF2hTERT non-tumoral fibroblasts determined by bisulfite sequencing (gray bars). Data are indicated as % ± SD of total CpG. Relative Sat2 cDNA/Sat2 gDNA ratio were calculated as follows: (Sat2 cDNA/ACTB cDNA)/(Sat2 gDNA/Alu gDNA) (black bars). Data are presented as % ± SEM of the maximal expression level measured in cell lines. (D) Linear regression of data from C. Black diamonds: sarcoma; gray dots: carcinoma; white dot: melanoma. (E) Normalized Sat2 RNA levels were measured in either HFF2 or HeLa cells after 1h of heat shock (HS) at 42°C followed by 1h of recovery at 37°C. Basal expression levels in U2OS, LB188 and SaOS-2 hypomethylated cancer cell lines are also shown. Data are presented as % ± SEM of the expression measured in HFF2.
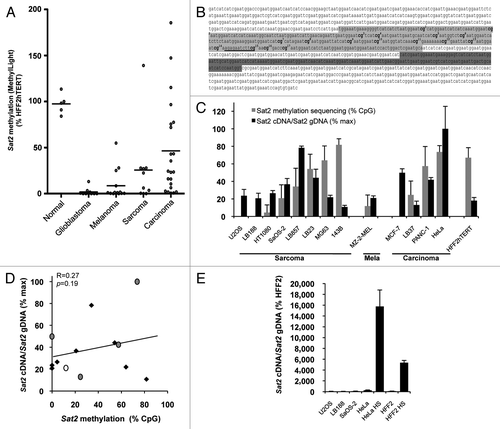
Figure 2.Sat2 hypomethylation in DNMT3B-deficient ICFhTERT fibroblasts does not exacerbate heat shock-induced Sat2 RNA expression. (A) Upper panel: schematic representation of the Sat2 fragment analyzed by sequencing. Positions of the 17 CpG sites and of the putative HSF1 binding site are indicated. Lower panel: Sat2 methylation level determined by sequencing of bisulfite-treated DNA molecules isolated from HFF2hTERT and ICFhTERT cells. White squares represent unmethylated CpG, black squares represent methylated CpG. (B) Relative Sat2 RNA levels in HFF2hTERT and ICFhTERT cells. Data are indicated as % ± SEM of the expression measured in HFF2hTERT. (C) Basal and HS-induced Sat2 and HSP70 RNA levels in HFF2hTERT and ICFhTERT. For HS, cells were incubated for 1h at 42°C followed by 1h of recovery at 37°C. Data are presented as % ± SEM of the basal expression measured in HFF2hTERT. (D) HFF2hTERT cells were incubated in the presence or absence (ctrl) of 5 μM 5-aza-2’-deoxycytidine (5-aza) for 1 to 5 d and relative expression levels of Sat2, MAGE-A1, MAGE-A4 and HSP70 were measured by qRT-PCR. Data are presented as % ± SEM of the expression measured in HFF2hTERT control cells.
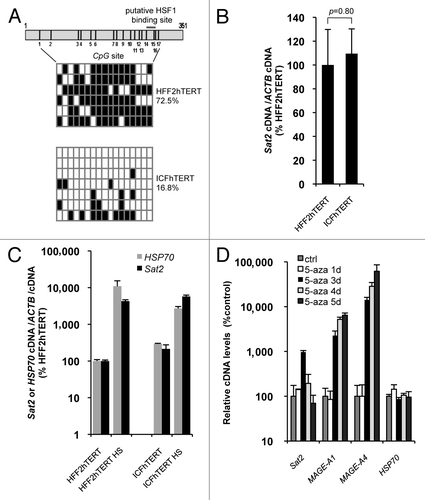
Figure 3. Hypomethylated Sat2 in ICFhTERT is characterized by increased density of H3K27me3 marks. (A) Density of H3K9me3 or H3K27me3 at Sat2 locus of HFF2hTERT and ICFhTERT determined by ChIP followed by qPCR. These marks were also quantified at GAPDH and TERRA (1q, 2q, 5p, 10q, 13q, 15q, 16p and 21q) promoters as controls. ChIP/input ratios were expressed as % ± SEM of the ratios obtained for Sat2 DNA in HFF2hTERT with either H3K9me3 or H3K27me3 antibody. (B) Western blot analysis of EZH2 and H3K27me3 after treatment of ICFhTERT cells with siRNAs against either Luciferase (siLuci) or EZH2 (siEZH2) for 72h. β-actin is shown as loading control. (C) Relative Sat2 RNA levels in ICFhTERT treated as in B and expressed as % ± SEM of Sat2 RNA in HFF2hTERT siLuci control cells.
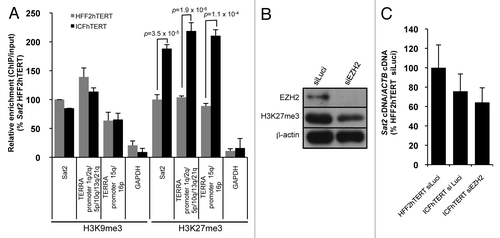
Figure 4.Sat2 RNA levels are correlated with HSP70 expression in melanoma tissues. (A) Relative Sat2 RNA levels measured in 6 primary melanoma tumors and 4 melanoma metastases. Data are presented as % ± SEM of the expression measured in HFF2. (B) Comparison between Sat2 and HSP70 expression levels in melanoma tissues from A. Both Sat2 and HSP70 cDNA levels were normalized to ACTB cDNA and expressed as % of the maximal value measured in the samples. (C) Melanoma tissues from A were classified into two groups according to the presence (+) or the absence (-) of MAGE-A2 or MAGE-A1 transcripts. Relative Sat2 RNA levels, as described in A, are indicated.
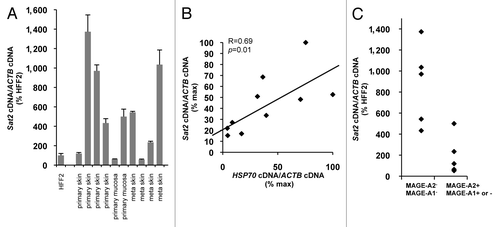
Figure 5. Heat shock and RasV12 oncogene induce Sat2 DNA demethylation. (A) Sat2 and HSP70 expression levels in HFF2hTERT cells subjected or not to HS for 1h at 42°C followed by either 1h or 4 d of recovery at 37°C. Data are given as % ± SEM of the expression levels measured in control cells. (B) Upper panel: schematic representation of the Sat2 fragment analyzed by sequencing. The 17 CpG dinucleotides and the putative HSF1 binding site are indicated. Lower panel: analysis of Sat2 methylation by sequencing of individual molecules amplified from bisulfite-treated DNA isolated from cells described in A. White squares represent unmethylated CpG, black squares represent methylated CpG. A Yates’ χ2 test was applied to assess the statistical differences in methylated CpG content with the control. (C) Relative Sat2 and HSF1 RNA levels in HFF2 fibroblasts 16 d after transduction with either empty pBabe or pBabe::RasV12 plasmid and expressed as % ± SEM of the expression in HFF2pBabe. (D) Sat2 methylation analysis of HFF2pBabe and HFF2pBabe::RasV12 determined 16 d after transduction. See B for details. (E) Average methylation level of CpG sites 1 to 10 (black bars) or 11 to 17 (gray bars) of Sat2 determined from B and D. A Yates’ χ2 test was applied to assess the statistical differences in methylated CpG content within CpG sites 11 to 17 in all tested conditions. (F) LINE-1 methylated CpG content determined by qMS-PCR on bisulfite-treated DNA prepared from samples described in B and D. [Methylated LINE-1 /(methylated LINE-1 + unmethylated LINE-1)] ratios were expressed as % ± SEM of LINE-1 methylation in control cells. (G) Analysis of D4Z4 and MAGE-A1 promoter methylation in HFF2pBabe and HFF2pBabe::RasV12 fibroblasts 16 d after transduction.
![Figure 5. Heat shock and RasV12 oncogene induce Sat2 DNA demethylation. (A) Sat2 and HSP70 expression levels in HFF2hTERT cells subjected or not to HS for 1h at 42°C followed by either 1h or 4 d of recovery at 37°C. Data are given as % ± SEM of the expression levels measured in control cells. (B) Upper panel: schematic representation of the Sat2 fragment analyzed by sequencing. The 17 CpG dinucleotides and the putative HSF1 binding site are indicated. Lower panel: analysis of Sat2 methylation by sequencing of individual molecules amplified from bisulfite-treated DNA isolated from cells described in A. White squares represent unmethylated CpG, black squares represent methylated CpG. A Yates’ χ2 test was applied to assess the statistical differences in methylated CpG content with the control. (C) Relative Sat2 and HSF1 RNA levels in HFF2 fibroblasts 16 d after transduction with either empty pBabe or pBabe::RasV12 plasmid and expressed as % ± SEM of the expression in HFF2pBabe. (D) Sat2 methylation analysis of HFF2pBabe and HFF2pBabe::RasV12 determined 16 d after transduction. See B for details. (E) Average methylation level of CpG sites 1 to 10 (black bars) or 11 to 17 (gray bars) of Sat2 determined from B and D. A Yates’ χ2 test was applied to assess the statistical differences in methylated CpG content within CpG sites 11 to 17 in all tested conditions. (F) LINE-1 methylated CpG content determined by qMS-PCR on bisulfite-treated DNA prepared from samples described in B and D. [Methylated LINE-1 /(methylated LINE-1 + unmethylated LINE-1)] ratios were expressed as % ± SEM of LINE-1 methylation in control cells. (G) Analysis of D4Z4 and MAGE-A1 promoter methylation in HFF2pBabe and HFF2pBabe::RasV12 fibroblasts 16 d after transduction.](/cms/asset/739d8c57-0cf9-42d7-81d1-3547c82d5f43/kepi_a_10921107_f0005.gif)
Figure 6. Heat shock pathway-dependent signature of Sat2 demethylation is also observed in cancer cell lines. (A) Left panel: methylation profile of Sat2, D4Z4 and MAGE-A1 promoter determined by sequencing of bisulfite-treated DNA molecules isolated from sarcoma (LB23, U2OS, LB188, HT1080), melanoma (LB45, MZ-2-MEL) and carcinoma (PANC-1, LB159) cell lines and from HFF2hTERT and HEKhTERT non-tumoral cell lines; right panel: LINE-1 methylation level determined by qMS-PCR on bisulfite-treated DNA with [methylated LINE-1 /(methylated LINE-1 + unmethylated LINE-1)] ratios expressed as % ± SEM (B) Relative methylation levels of CpG sites 1–10 or 11–17 of Sat2 in LB23, LB45, PANC-1 and LB159 cancer cell lines compared with the methylated CpG content of Sat2 DNA from HFF2hTERT and HEKhTERT control cells together. Yates’ χ2p-values are indicated.
![Figure 6. Heat shock pathway-dependent signature of Sat2 demethylation is also observed in cancer cell lines. (A) Left panel: methylation profile of Sat2, D4Z4 and MAGE-A1 promoter determined by sequencing of bisulfite-treated DNA molecules isolated from sarcoma (LB23, U2OS, LB188, HT1080), melanoma (LB45, MZ-2-MEL) and carcinoma (PANC-1, LB159) cell lines and from HFF2hTERT and HEKhTERT non-tumoral cell lines; right panel: LINE-1 methylation level determined by qMS-PCR on bisulfite-treated DNA with [methylated LINE-1 /(methylated LINE-1 + unmethylated LINE-1)] ratios expressed as % ± SEM (B) Relative methylation levels of CpG sites 1–10 or 11–17 of Sat2 in LB23, LB45, PANC-1 and LB159 cancer cell lines compared with the methylated CpG content of Sat2 DNA from HFF2hTERT and HEKhTERT control cells together. Yates’ χ2p-values are indicated.](/cms/asset/28ef12b3-1935-4530-9240-26ac3c0583b5/kepi_a_10921107_f0006.gif)