Figures & data
Figure 1. Mutations nrd1, nrd2–1 and nrd2–2 release RdTGS and RdDM of a ProNOS-NPTII reporter gene. (A) Transgene system: The SILENCER (H) transgene contains an inverted repeat (IR) of the NOPALINE SYNTHASE promoter (ProNOS) sequence under control of the strong constitutive cauliflower mosaic virus 35S promoter (Pro35S). Transcripts of the ProNOS-spacer-SONorP structure can fold to form double-stranded RNA with ProNOS-homology, which is then processed to short interfering (si)RNAs. These siRNAs serve as a signal for in trans DNA methylation and transcriptional silencing of a ProNOS copy that controls transcription of a NEOMYCIN PHOSPHOTRANSFERASE II (NPTII) (conferring kanamycin resistance if expressed) in the unlinked TARGET (K) transgene. In addition, the SILENCER contains a HYGROMYCIN PHOSPHOTRANSFERASE (HPT) gene conferring hygromycin resistance and the TARGET a GUS reporter gene (not shown). (B) Test for kanamycin resistance on medium containing 200 mg/l kanamycin. (C) Quantification of NPTII protein by ELISA: NPTII protein amouts in relation to total soluble protein were measured in extracts from leaves of 8-week-old plants. Results are displayed relative to the mean value for un-silenced expression in (K/K;-/-) plants (set to 1). Per genotype, five individual plants were tested. Column hights represent mean values; error bars represent standard deviations. (D) TARGET ProNOS cytosine methylation was determined by quantitative PCR after cleavage of genomic DNA from 8-week-old plants with methylation-sensitive restriction enzymes (C in recognition sequence underlined: methylation of cytosine blocks cleavage according to REBASE http://rebase.neb.com/rebase/rebase.html) Psp1406I (olive, symmetric CG context: AACGTT), NheI (blue, CHG and CHH context: GCTAGC), Alw26I (orange, asymmetric CHH context: GTCTC, GAGAC) and NcoI (yellow, asymmetric CHH context, control outside of the methylated region: CCATGG). Per genotype, five individual plants were tested. Results are displayed relative to the mean value for input DNA (set to 1). Column highs represent mean values; error bars represent standard deviations.
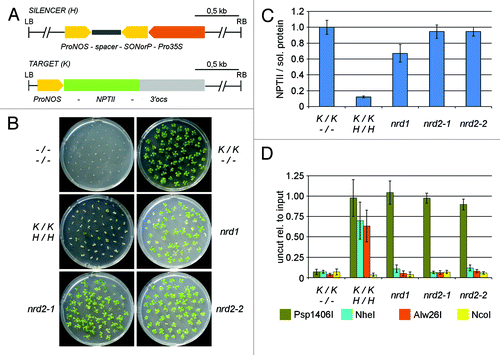
Figure 2. Map-based cloning of nrd1. (A) Physical map indicating markers and recombination events (numbers in parentheses, of 234 chromosomes in total) used to delineate the position of nrd1 on the lower arm of chromosome 3. (B) Positions of the nucleotide (top) and related amino acid change (bottom) in nrd1 in the IDN2 gene model (according to TAIR 10). (C) Complementation of nrd1 by transgenic IDN2. Seeds were germinated on medium containing 200 mg/l kanamycin. Approx. 75% of T2 progeny obtained by self-pollination of (single locus) nrd1 + IDN2 T1 transformants inherit a transgenic functional IDN2 and thus are sensitive to kanamycin due to re-establishment of RdTGS of the ProNOS-NPTII gene.
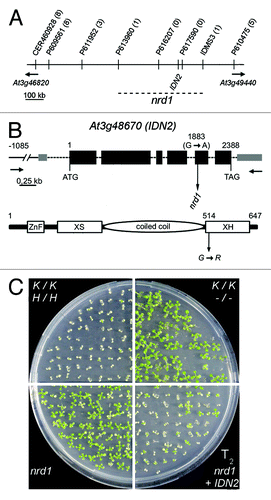
Figure 3. Map-based cloning of nrd2–1 and nrd2–2. (A) Physical map indicating markers and recombination events (numbers in parentheses separated by semicolon, of 172 chromosomes in total for nrd2–1 and 134 for nrd2–2, respectively) used to delineate the position of nrd2 on the upper arm of chromosome 3. (B) Positions of the nucleotide (top) and related amino acid change (bottom) in nrd2–1 and nrd2–2 in the NRPD2a/NRPE2a gene model (according to TAIR10).
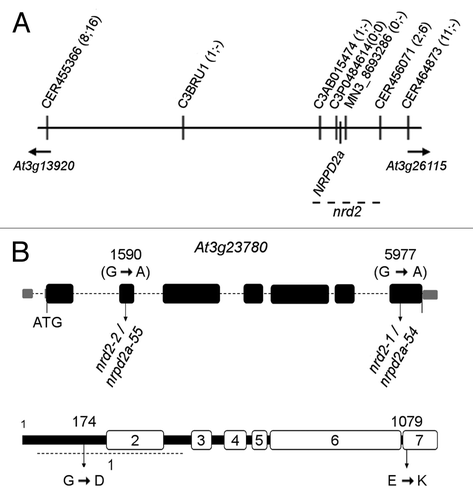
Figure 4. Detailed TARGET ProNOS DNA methylation analysis in idn2–8, nrpd2a-54 and nrpd2a-55. DNA methylation patterns in the ProNOS of the ProNOS-NPTII reporter gene were analyzed in detail by bisulfite sequencing. (A) Cumulative methylation levels at all cytosines in the analyzed region (gray columns), cytosines in CG context (black columns), CHG context (blue columns; H stands for A, C or T) and CHH context (red columns). (B) Methylation levels at individual cytosines in CG context (black columns), CHG context (blue columns) and CHH context (red columns). A black arrowhead marks the ProNOS transcription start site. Numbers of clones sequenced per target and genotype were: 15 (K/K −/−), 19 (K/K;H/H), 20 (idn2–8), 15 (nrpd2a-54), 21 (nrpd2a-55)
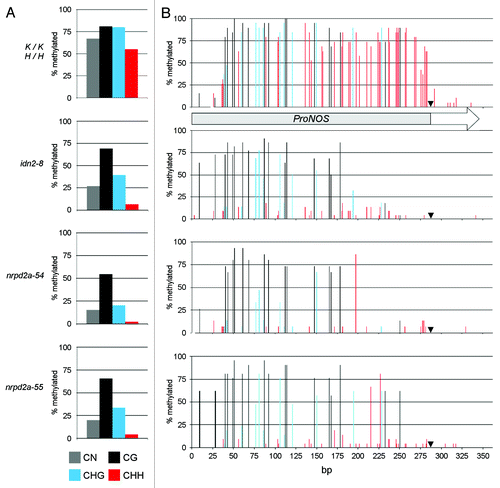
Figure 5. Detailed DNA methylation analysis at endogenous AtSN1, MEA-ISR, AtMu1 and AtCOPIA4 sequences in idn2–8, nrpd2a-54 and nrpd2a-55. DNA methylation patterns of endogenous sequences (A) AtSN1, (B) AtMU1, (C) MEA-ISR and (D) AtCOPIA4 were analyzed in detail by bisulfite sequencing in non-mutagenized control plants (black columns), idn2–8 (dark gray columns), nrpd2a-54 (light gray columns) and nrpd2a-55 (white columns). A minimum of 12 clones was sequenced per target and genotype. Exact numbers are indicated in Figures S8–10.
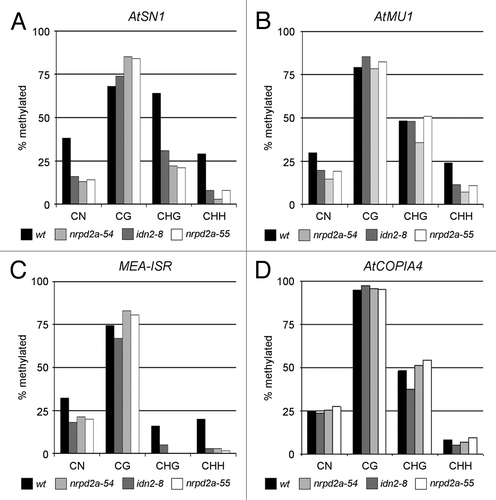
Figure 6.ProNOS-derived and endogenous siRNAs in idn2–8 and nrpd2a-55. Northern blot for siRNA derived from transcription of ProNOS IR in the SILENCER. (A) Blots were hybridized with a RNA probe specific for sense ProNOS siRNAs. (B) Equal loading was confirmed by re-hybridization with miR167-specific probe after stripping.
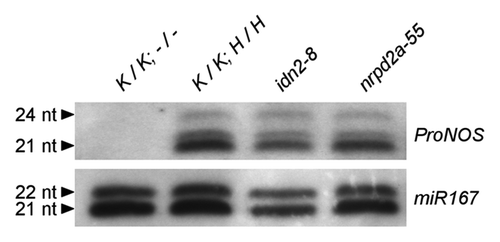
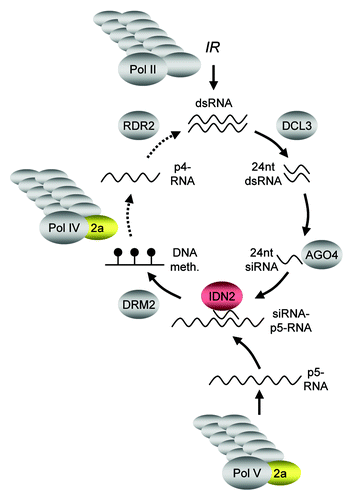
Figure 7. Genetic model of RdDM. The core pathway leading to RdDM is initiated by the production of single stranded RNA from target sequences by multi-subunit RNAP IV. The resulting p4-RNA then serves as substrate of RDR2, which synthesizes a complementary strand to generate dsRNA. This dsRNA is then processed by DCL3 into 24 nt dsRNA fragments and single strands of 24 nt short interfering (si)RNA are incorporated mainly into AGO4. Multi-subunit RNAP-V is thought to transcribe RdDM target loci, with the resulting p5-RNA serving as scaffold to attract the siRNA-AGO4 complexes, which in turn guide DRM2 to the genomic loci to be methylated de novo. Transcription of an inverted repeat (IR) by multi-subunit RNAP II provides a shortcut in the pathway, as dsRNA as a substrate for DCL3 action is produced independently of RNAP IV and RDR2 (solid arrows). NRPD2a/NRPE2a (golden) is a subunit common to RNAP IV and RNAP V, but is not required for RNAP II function. IDN2 (red) has a role downstream of siRNA formation, possibly by stabilizing a siRNA-p5-RNA complex.