Figures & data
Figure 1. Assessment of microarray data quality by ArrayQualityMetrics package of Bioconductor and normalization of biological replicates. A. Scatterplot and Spearmann correlation of raw intensities between two biological replicates. The ArrayQualityMetrics tool allowed generation of correlation co-efficiency value between ChIP-on-chip replicates CC = 0.783 (0, no correlation; 1, perfect correlation). (B) Histogram of mean signal ratios of H4K12ac chromatin-immunoprecipitated DNA vs. random-sheared total genomic DNA (Input). The distinct tail at the right-hand end corresponds to DNA fragments enriched by H4K12ac. Tiled oligos that displayed the top 3% ratios are located to the right of the red bar. (C) The highest peak obtained after averaging of 2 replicates as calculated by log ratios between input material and immunoprecipitated sample.
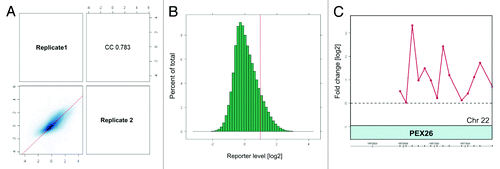
Figure 2. Distribution of H4K12ac peaks (shown as bars) along chromosome 1 from one healthy donor aligned alongside Ensembl gene density profile and ideograms of chromosome 1. The positions of the H4K12ac peaks on were plotted against their enrichment, which was calculated from the log2-ratio of input signal-Cy3 for the total chromatin (control) and IP-probe–Cy5. False discovery rate (FDR) predicts significance of peak enrichment. (Red bars; p < 0.001, yellow bars; p < 0.05, gray bars p > 0.05). Higher density of binding sites was recognized in telomeric sequences and region of high gene density on chromosome 1. Table summarizes the frequency of binding of all human chromosomes.
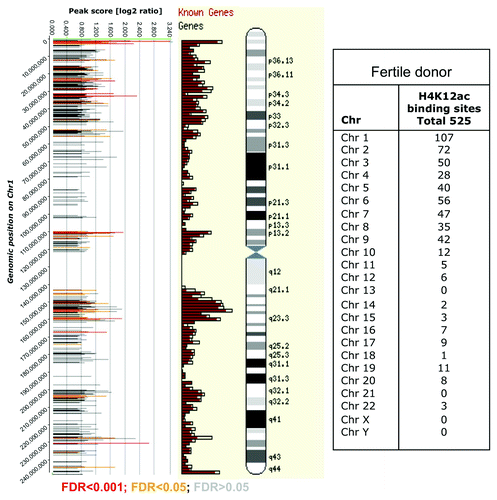
Figure 3. H4K12ac-associated promoters involved in reproductive process (A), embryonic development (B) and chromatin organization (C). Spermatogenesis / fertility-related and developmentally important gene promoters selected according to gene ontology classification available as a web tool (http://david.abcc.ncifcrf.gov/).
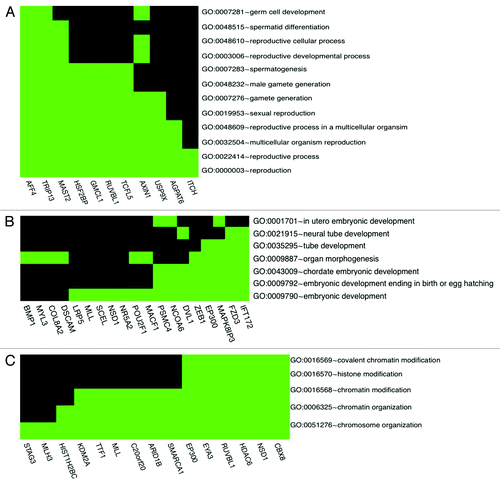
Figure 4. Validation of ten enriched promoters for H4K12ac in spermatozoa. Results from ChIP assay with anti-H4K12ac antibodies, with anti-protamine in immunoprecipitated DNA of 3 fertile donors, in combination with real time PCR. Unmodified H3 was used as positive and IgG as negative control.
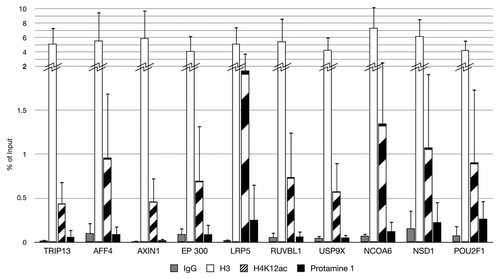
Figure 5. Distribution of H4K12ac binding sites relative to transcription start sites (TSS) of the spermatozoal genome. Histogram generated using ChIPpeakAnno package of R software shows that H4K12ac interacts with regions symmetrically distributed around TSS. The highest binding frequency could be observed 2 kb upstream and 2 kb downstream from TSS.
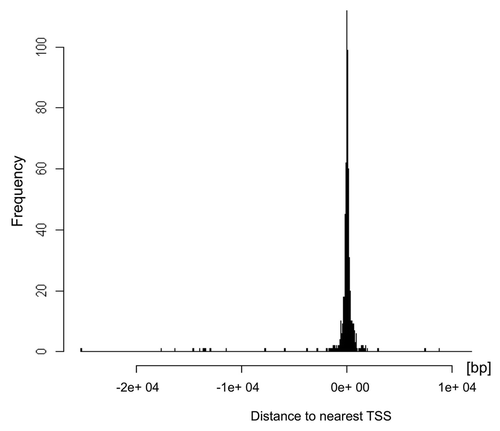
Figure 6. Venn diagram demonstrating co-localization of H4K12ac with methylated histones: H3K27me3, and H3K4me3 (A), and H3K9ac (B) within promoters of human sperm. Analysis was performed with data published by Hammoud et al.Citation14 and ENCODE CHIP-on Chip with H3K9ac from our previous study.Citation13
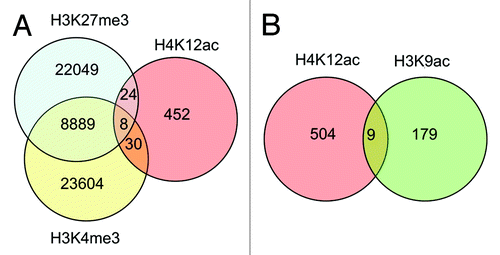
Figure 7. mRNA expression levels of H4K12ac-associated genes in spermatozoa. The plot demonstrates the distribution (as relative frequencies, ordinate) of microarray derived relative mRNA expression levels (abscissa) in the group of H4K12ac-associated genes (red bars) compared with the group of all 20,400 genes (gray bars) from fertile donors. The list of H4K12ac-associated promoters with corresponding mRNA transcripts in sperm of healthy donors are shown in Figure S2.
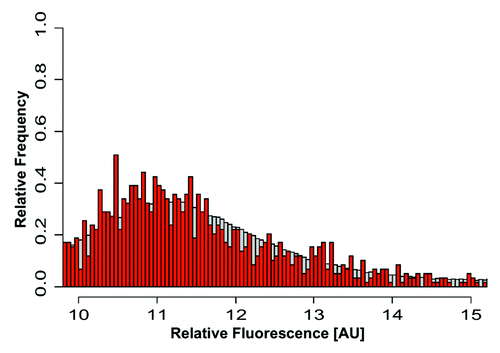
Figure 8. Distribution of H4K12ac in human sperm nucleus and during mouse early embryo development. Co-localization of H4K12ac (A) and protamine-1 (B) visualized by immunofluorescence using FITC (green) and Cy3 (red) conjugated antibodies (merge) (C). DAPI was used for DNA staining (blue) (D). Immunolabeling of H4K12ac was detected in the post-acrosomal region of the sperm head, while red staining of protamine-1 was spread over the whole nucleus. Immunofluorescent labeling of H4K12ac in early stages of pronuclei establishment (E–H); Pronuclei fusion (I–L); H4K12ac staining (green). Male pronucleus, shown by arrow, gives strong positive signal compares to a smaller female pronucleus shown by asterisk; DNA stained by DAPI (blue) (B,F,); H4K12ac and DAPI (C,G); H4K12ac, DAPI and DIC (D,H). Scale bars represent 10 µm. Photos are merged for each displayed color labeling from several sequenced slices taken by a Leica confocal microscope.
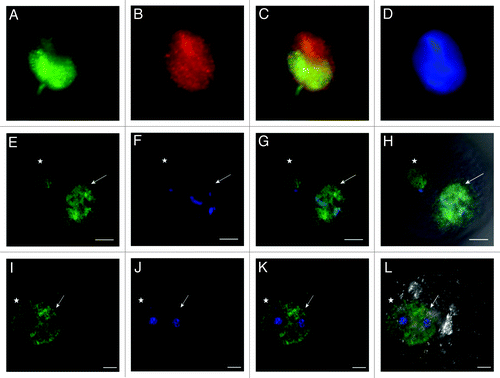
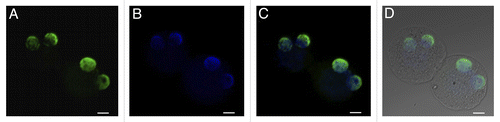
Figure 9. Distribution of H4K12ac in parthenogenetically activated oocytes. Maternal pronuclei after parthenogenetic activation display identical strong positive signal. H4K12ac (A), DAPI staining (B); H4K12ac with DAPI (C); H4K12ac, DAPI and DIC (D). Scale bars represent 20 µm.