Figures & data
Figure 1. DNA methylation in MACS purified human leukocyte samples at immune cell-specific DMRs. (A) Illumina Infinium HumanMethylation27 data for top 100 putative T cell-specific DMRs identified from LME model. Beta values are scaled (zero centered) by gene locus. (B) MethyLight data for FOXP3 and CD3Z gene regions illustrating immune lineage-specific DNA methylation patterns. MACS purified cell type abbreviations: B, B lymphocytes; Gran, granulocytes; Neut, neutrophils; Mono, monocytes; NK, CD56+ natural killer cells; CD16+NK, CD16+CD56dim natural killer cells; CD16-NK, CD16-CD56bright natural killer cells; NKT, CD3+CD56+ natural killer T-cells; t, CD3+ T lymphocytes; Tc, CD3+CD8+ T lymphocytes (cytotoxic T-cells); Th, CD3+CD4+ T lymphocytes (helper T-cells); Treg, CD3+CD4+CD25+FOXP3+ regulatory T-cells
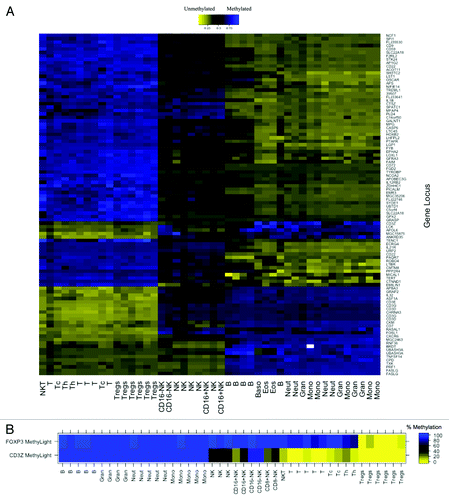
Figure 2. CD3+ T-cell detection by qMSP. (A) Schematic showing methylation specific primers and probe targeting six CpGs (lollipops) in a region of the CD3Z gene identified as demethylated in CD3+ T-cells. (B) Real time PCR Ct values decrease linearly with 10-fold increase in bisulfite converted CD3+ T-cell DNA concentration. Bisulfite converted universal methylated DNA was used to keep total amount of DNA in all samples constant. At least five replicates of each sample are plotted. (C) Evaluation of CD3+ T-cell level by flow cytometry is highly correlated with T-cell quantification by CD3Z qMSP in whole blood specimens from glioma patients and healthy donors. (D) CD3+ T-cell count by imunohistochemical staining correlates with T-cell quantification by CD3Z qMSP in excised tumors across histological subtypes. Pearson correlations and F-test p values are shown in B‒D.
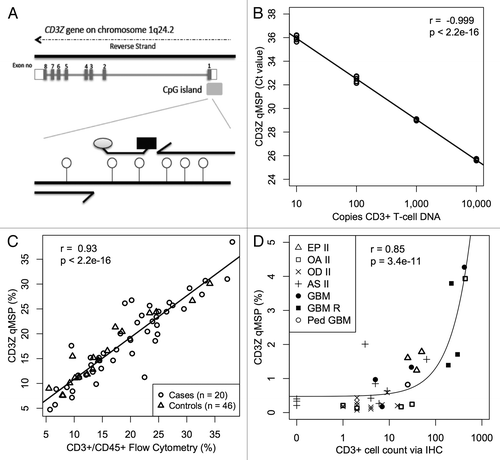
Table 1. Summary of qMSP measurements for all samples (n = 285)
Figure 3. T-cells and Tregs in the peripheral blood of glioma patients and healthy donors. (A) Case control comparison of T-cells measured via CD3Z demethylation assay. (B) Case control comparison of Tregs measured via FOXP3 demethylation assay. (C) Case control comparison of Treg percent of T-cells determined by the ratio of FOXP3/CD3Z demethylation. In each panel, the displayed p value is from a Wilcoxon rank sum test between non-diseased control bloods and GBM patient bloods. Each data point represents the average of all replicate qMSP measurements for a single individual. Box plots superimposed on the data points cover the 2nd and 3rd quartile range with a line drawn at the median value, and whiskers that extend to the data point that is no more than 1.5 times the length of the box away from the box. The notches surrounding each median line extend to ± 1.58 IQR/sqrt(n), such that if the notches from two boxplots do not overlap there is strong evidence for a significant difference in the two medians.
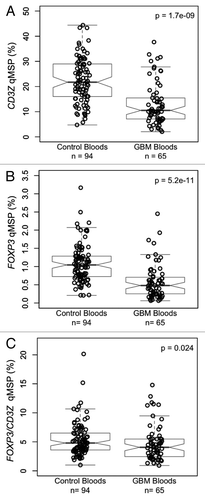
Figure 4. Association between cigarette smoking and peripheral blood T-cells and Tregs in glioma patients and healthy donors. (A) Comparison of peripheral blood T-cell levels, determined by CD3Z demethylation, among never, former and current cigarette smokers stratified by glioma case status. (B) Comparison of peripheral blood Treg levels, determined by FOXP3 demethylation, among never, former and current cigarette smokers stratified by glioma case status. (C) Comparison of peripheral blood Treg percent of T-cells, determined by ratio of FOXP3 to CD3Z demethylation, among never, former and current cigarette smokers stratified by glioma case status. In each panel the displayed p-values are from Wilcoxon rank sum tests between the two groups indicated. Each data point represents the average of all replicate qMSP measurements for a single individual. Box plots superimposed on the data points cover the 2nd and 3rd quartile range with a line drawn at the median value, and whiskers that extend to 1.5 times the length of the box.
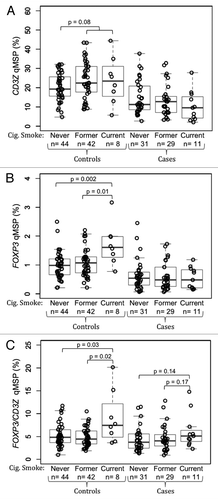
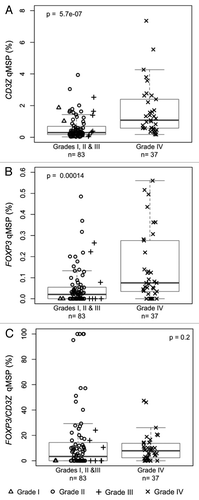
Figure 5. Levels of T-cell and Treg infiltrates in excised glioma tissue. (A) T-cell levels, determined by CD3Z demethylation, in solid glioma samples stratified by tumor grade. (B) Treg levels, determined by FOXP3 demethylation, in solid glioma samples stratified by tumor grade. (C) Treg percent of T-cells, determined by ratio of FOXP3 to CD3Z demethylation, in solid glioma samples stratified by tumor grade. In each panel the displayed p-value is from a Wilcoxon rank sum test between lower grade tumors (Grades I, II and III) and Grade IV tumors. Each data point represents the average of all replicate qMSP measurements for a single individual. Box plots superimposed on the data points cover the 2nd and 3rd quartile range with a line drawn at the median value, and whiskers that extend to 1.5 times the length of the box.