Figures & data
Figure 1. Differential expression of NKG2D gene in T and NK cell lines and lymphocyte subsets is correlated with DNA methylation pattern.(A) NKG2D transcription levels were determined by qRT-PCR in Jurkat, HUT78 and NKL cell lines, and isolated CD4+ T, CD8+ T and NK cells from healthy donors. Histograms represent the relative expression of NKG2D gene normalized with respect to the housekeeping gene GAPDH. Data are presented as the mean ± SD of five independent experiments. (B) Graphic representation of the NKG2D gene. Arrow: translation start site; filled bars: distribution and position of CpG sites; horizontal black lines: regions amplified with specific primers shown in . The methylation pattern of each cell type was analyzed by bisulfite sequencing. Rows: ten individual sequenced clones; columns: CpG sites; closed squares: methylated CpG sites; open squares: unmethylated CpG sites. The right-hand graphs show the total percentage methylation for each sample.
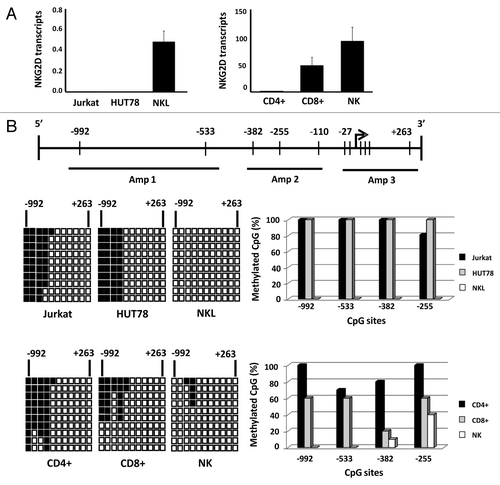
Figure 2. Effect of DNMT inhibitors (5azaC and DAC) in NKG2D expression in Jurkat and CD4+ T cells. (A) Jurkat cells were treated with 5azaC (1–5 μM) and DAC (60–80 μM) for 24, 48 and 72 h and NKG2D transcripts were quantified by qRT-PCR using specific primers (). Histograms show the fold increase of NKG2D expression after treatment relative to untreated cells. GAPDH was used as a constitutive control and data are presented as the mean ± SD of three independent experiments. The right panel shows representative histograms of NKG2D expression on the cell surface of Jurkat cells after DNMT inhibitors treatment for 72 h. (B) NKG2D transcription levels were determined by qRT-PCR in CD4+ T lymphocytes from healthy donors. Isolated CD4+ cells were previously activated with anti-CD3 and anti-CD28 mAb over four days, and further cultured in the absence or presence of DAC (1 μM) and 5azaC (5 μM) for 72 h. The histogram shows the fold increase of NKG2D expression after treatment relative to untreated cells. GAPDH was used as a constitutive control for qRT-PCR. Data are presented as the mean ± SD of three independent experiments. The right panel shows representative dot plots of the induced NKG2D expression on CD4+ T cells after DNMT inhibitor treatment. The number indicates the percentage of NKG2D-positive CD4+ T cells. *p < 0.05. Significance is shown as the difference between treated and untreated Jurkat or CD4+ T cells.
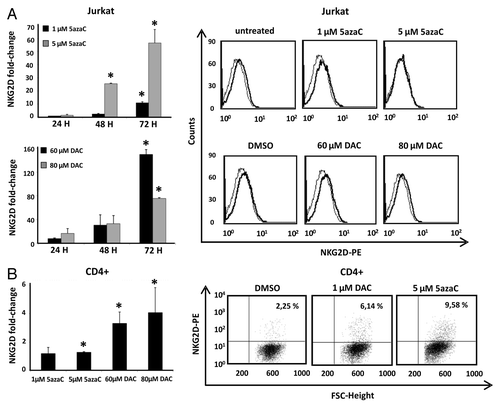
Figure 3. Enrichment of active and repressive histone modifications in NKG2D gene. Levels of histone modifications in NKG2D gene were measured by chromatin immunoprecipitation (ChIP) assay using antibodies specific for AcH4, AcH3, H3K9Ac, H3K4me, H3K27me3, H3K9 me2 in the Jurkat, HUT78 and NKL cell lines (A) and in CD4+, CD8+ T lymphocytes and NK cells (B). Normal rabbit IgG was used as a negative control. Enrichment of NKG2D-specific DNA sequences was measured by quantitative PCR with specific primers (). Two consecutive regions in the NKG2D gene were amplified and the average of both products is shown. Results are represented as % INPUT = 2 exp [Ct (BOUND) – (Ct (UNBOUND) - log2 (UNBOUND DILUTION FACTOR))] × 100. Data are shown from two independent experiments and each qRT-PCR was performed in triplicate.*p < 0.05 for comparisons of Jurkat and NKL cells, and of CD4+ T cells and NK cells. #p < 0.05 for comparisons of HUT78 and NKL cells, and of CD4+ T with CD8+ T cells.
![Figure 3. Enrichment of active and repressive histone modifications in NKG2D gene. Levels of histone modifications in NKG2D gene were measured by chromatin immunoprecipitation (ChIP) assay using antibodies specific for AcH4, AcH3, H3K9Ac, H3K4me, H3K27me3, H3K9 me2 in the Jurkat, HUT78 and NKL cell lines (A) and in CD4+, CD8+ T lymphocytes and NK cells (B). Normal rabbit IgG was used as a negative control. Enrichment of NKG2D-specific DNA sequences was measured by quantitative PCR with specific primers (Table 1). Two consecutive regions in the NKG2D gene were amplified and the average of both products is shown. Results are represented as % INPUT = 2 exp [Ct (BOUND) – (Ct (UNBOUND) - log2 (UNBOUND DILUTION FACTOR))] × 100. Data are shown from two independent experiments and each qRT-PCR was performed in triplicate.*p < 0.05 for comparisons of Jurkat and NKL cells, and of CD4+ T cells and NK cells. #p < 0.05 for comparisons of HUT78 and NKL cells, and of CD4+ T with CD8+ T cells.](/cms/asset/0a0fa1d6-7ce5-412c-a3bd-b69e7fc3fcc8/kepi_a_10923115_f0003.gif)
Table 1. PCR and qRT-PCR primers
Figure 4. Effect of HDAC and HAT inhibitors in NKG2D expression in Jurkat and NKL cell lines. (A) NKG2D expression was measured in the Jurkat cell line after treatment with VPA (0.5–1.0 mM) in the absence or presence of IL-15 (50 ng/ml) cytokine. Cells were harvested after four days and NKG2D transcripts were quantified by qRT-PCR using specific primers (). The histogram shows the fold increase in NKG2D expression after treatment relative to untreated cells. GAPDH was used as a constitutive control for qRT-PCR. Data are represented as mean ± SD of three independent experiments *p < 0.05. Significance is shown as the difference between untreated and treated Jurkat-cells. (B) NKG2D expression in NKL cells was analyzed before and after treatment with 4 μM curcumin for 24 h by flow cytometry. The histogram is representative of five independent experiments. (C) NKG2D and DAP10 transcripts were quantified by qRT-PCR in untreated and curcumin-treated NKL cells (4 µM, 24 h). GAPDH was used as a control and the results are represented as the mean ± SD of three independent experiments. (D) The level of H3K9Ac in the NKG2D gene was analyzed in untreated and curcumin-treated NKL cells by ChIP assay. Normal rabbit IgG was used as a negative control. Data are expressed as the % INPUT from three independent experiments and qRT-PCR was performed in triplicate. *p < 0.05
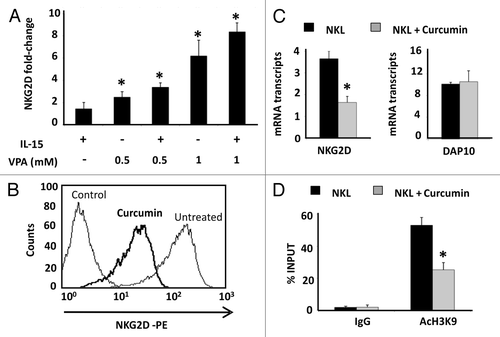
Figure 5. Downregulation of NKG2D expression in CD8+ T lymphocytes and NK cells after curcumin treatment. NKG2D expression on the cell surface of CD8+ T cells (A) and NK cells (B) was assayed before and after treatment with curcumin (1–4 μM) for 24 h by flow cytometry. Left-hand panels show a representative histogram after treatment with 4 μM curcumin for 24 h; on the right, box-plots indicate NKG2D expression in CD8+ T lymphocytes and NK cells from ten healthy donors before and after curcumin treatment (1–4 μM). Data are expressed as the Mean Fluorescence Intensity (MFI) of NKG2D expression. *p < 0.05.
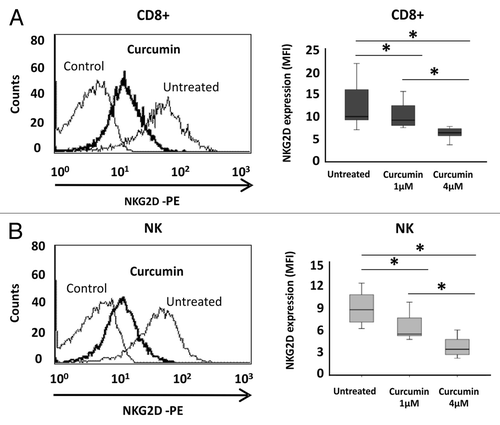
Figure 6. Curcumin suppresses NKG2D-mediated NKL cell line cytotoxicity. (A) NKL cells were cultured with C1R cells and C1R-MICA*004 transfectants at the indicated effector:target (E:T) ratios for 4 h. Cytotoxicity was assayed by flow cytometry after PKH67/7AAD staining. For blocking assays, NKL cells were incubated beforehand with anti-NKG2D blocking monoclonal antibody (10 μg/ml) for 30 min. Data are shown as means ± SD of two independent experiments in triplicate. (B) NKL cells were incubated in the absence or presence of curcumin (4 µM for 24 h) and cytolytic activity against C1R and C1R-MICA*004 cells was assessed at the indicated effector: target (E:T) ratios. The upper figure shows a representative experiment and the lower histogram represents means ± SD of three independent experiments performed in triplicate. *p < 0.05 comparison between untreated and curcumin- or anti-NKG2D-treated NKL cells against C1R-MICA*004 transfectants. #p < 0.05 C1R-MICA*004 compared with C1R cells.
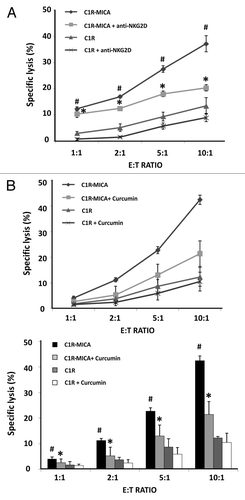
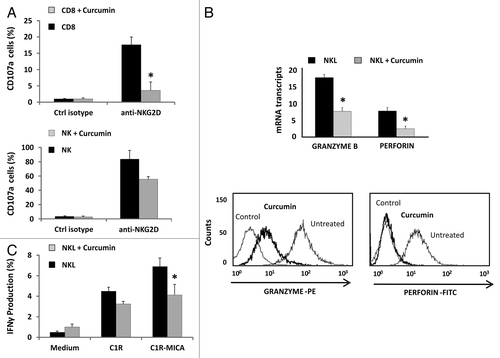
Figure 7. Curcumin modifies the cytolytic machinery in CD8+ T lymphocytes, NK and NKL cells. (A) Frequency of CD107a+ degranulation by T CD8+ and NK cells after stimulation with anti-NKG2D mAb. Untreated or curcumin-treated (4 µM) CD8+ and NK cells were cultured for 6 h in the presence of activating anti-NKG2D mAb or isotype control, and expression of membrane CD107a was measured by flow cytometry. Data represent the mean ± SD values from triplicate experiments. (B) Granzyme B and perforin expression was analyzed in the NKL cell line before and after treatment with 4 µM of curcumin for 24 h by qRT-PCR and flow cytometry. GAPDH was used as a control and results are represented as the mean ± SD of three independent experiments. Histograms are representative of an experiment for intracellular staining of granzyme B and perforin. (C) Frequency of IFN-γ production by untreated or 4 µM curcumin-treated NKL cells after coculture for 6 h with C1R or C1R-MICA cells at E: T ratio 1:1. Data represent mean ± SD values from triplicate experiments.*p < 0.05.