Figures & data
Figure 1. Phylogenetic tree of SET1/MLL family proteins. Relationships of planarian SET1/MLL proteins (highlighted in red) to those of other species based on Neighbor-joining analysis of their SET domains. Numbers represent the percentage of Bootstrap replicates that include that node. The predicted domain structure of each protein is diagramed to the right. The members of the larger family fall into four sub-groups indicated by brackets. Sequences were selected from Homo sapiens (HUMAN), Mus musculus (MOUSE), Caenorhabditis elegans (CAEEL), Schizosaccharomyces pombe (SCHPO), Saccharomyces cerevisiae (YEAST), Drosophila melanogaster (DROME) and Schmidtea mediterranea (SMED).

Figure 2. Expression of planarian set1/mll family genes. Whole mount in situ hybridization showing the mRNA expression patterns of genes in the S. mediterranea set1/mll family. The lower worm of each pair was treated with 60 Gy of γ−irradiation three days prior to fixation. mes, mesenchyme; cg, cephalic ganglia; int, intestine. Animals are shown ventral side up with anterior to the left. Scale bars = 0.5 mm.
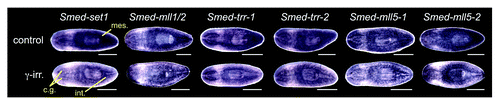
Figure 3.set1/mll family knockdown phenotypes. Planarians were fed bacterially expressed double-stranded RNA targeting each set1/mll gene and then transected anterior to the pharynx and observed during regeneration. gfp(RNAi) serves as a negative control. Animals in the left panel were fed three times over one and a half weeks and imaged after six days of regeneration. The animals in the right panel were fed six times over three weeks and imaged after seven days of regeneration. Yellow dashed lines mark the plane of amputation. White triangles indicate reduced blastema growth, green arrowheads indicate normal photoreceptors, and magenta arrows indicate lack of or underdeveloped photoreceptors. Anterior is up. Scale bars = 0.5 mm.
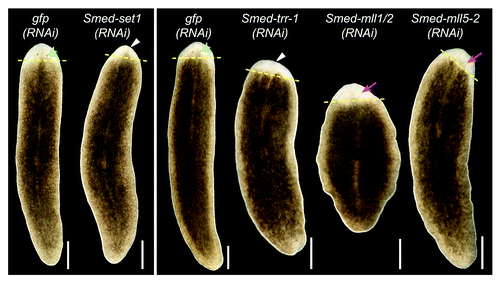
Figure 4. Effect of set1/mll knockdown on the mitotic stem cell population. (A) Anti-phospho-Histone H3 (Ser10) staining following RNAi against set1/mll family genes. Animals in the upper panel received four RNAi feedings over the course of two weeks, and worms in the lower panel received nine feedings over 4.5 weeks. Samples were fixed five days after the final feeding. Scale bars = 0.5 mm. (B) Quantitation of the number of phospho-Histone H3-positive cells in each group shown in A. Error bars represent standard error of the mean. Results significantly different from gfp(RNAi) control are marked by ** (p value < 0.01, Student’s t-test).
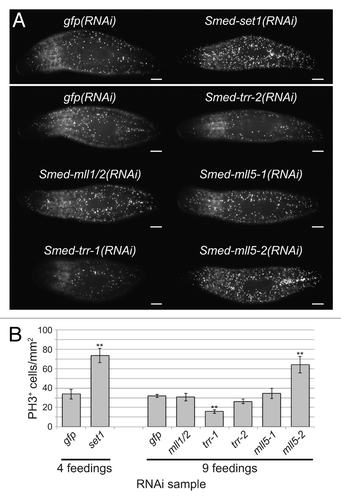
Figure 5. RNAi of Smed-set1 results in loss of stem cells. (A) In situ hybridization to markers of stem cell and progeny cell populations following knockdown of Smed-set1. Worms were fed dsRNA against Smed-set1 or gfp (negative control) four times over two weeks and then fixed and stained for markers of stem cells and their descendants. Scale bars = 0.5 mm. (B) Flow cytometric analysis of Smed-set1(RNAi) worms. Worms from the same RNAi knockdown as in A were dissociated and analyzed by flow cytometry. Some gfp(RNAi) worms were treated with 100 Gy γ-irradiation four days prior to dissociation to aid in identification of the cycling cells. (C) Summary of change in the percentage of cells in X1 from two replicates of the flow cytometric analysis shown in B. Error bars represent standard error of the mean. The difference between the two groups is statistically significant (p value < 0.01, Student’s t-test).
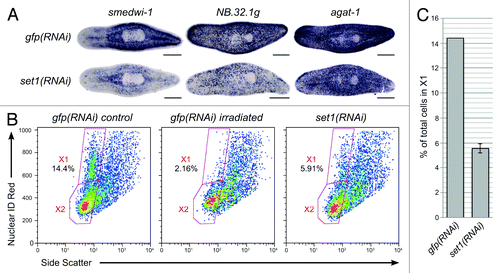
Figure 6. Identification of planarian COMPASS proteins. (A) Human COMPASS and COMPASS-like complexes. SET1/MLL family members (shown in gray) act in complex with other proteins. Core subunits common to all COMPASS and COMPASS-like complexes are shown in dark blue. Complex-specific subunits are color-coded by complex: red, SET1/COMPASS complex; orange, SET1/COMPASS and MLL1/2 COMPASS-like complexes; green, MLL1/2 COMPASS-like complex; light blue, MLL3/4 COMPASS-like complex. (B) S. mediterranea homologs of COMPASS proteins. Color-coding matches that in A. e-values are for reciprocal protein blast of the predicted full-length planarian protein against the top human hit in the NCBI non-redundant protein sequence database.
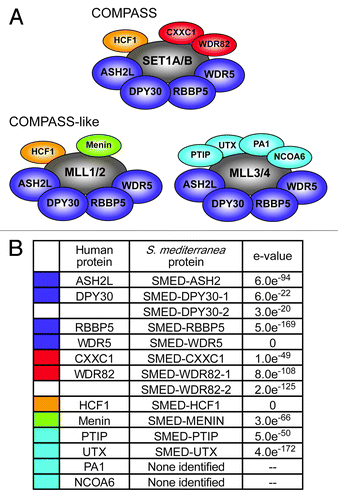
Figure 7. Expression of planarian COMPASS genes. Whole mount in situ hybridization to homologs of COMPASS and COMPASS-like complex members. Colored bars above the gene names indicate complex membership as in . The lower worm of each pair was treated with 60 Gy γ-irradiation three days prior to fixation. Mes, mesenchyme; cg, cephalic ganglia; int, intestine. Animals are shown ventral side up with the anterior to the left. Scale bars = 0.5 mm.
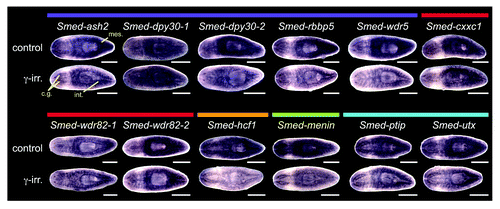
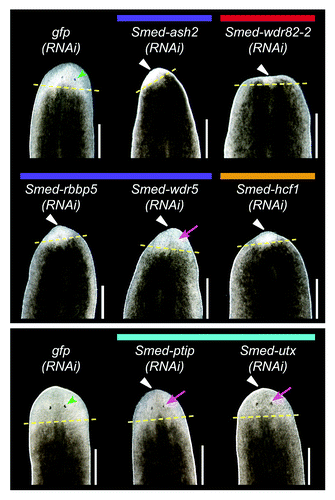
Figure 8. RNAi knockdown of COMPASS complex genes. Planarians were fed dsRNA targeting the indicated gene six times over three weeks and then amputated pre-pharyngeally to assay regeneration. gfp(RNAi) served as a negative control. The worms in the upper and lower panels were imaged after six or ten days of regeneration, respectively. Yellow dashed lines mark the plane of amputation. White triangles indicate reduced blastema formation, green arrowheads indicate normal photoreceptors, and magenta arrows indicate underdeveloped photoreceptors. Scale bars = 0.5 mm.