Figures & data
Figure 1. Cell culture condition for placental trophoblast samples. Cell cultures were plated and maintained in a culture support center at 37°C in a 5% carbon dioxide/air atmosphere (20% oxygen, standard conditions). After 4 h to allow attachment, half of the cells from each placenta were continued in standard conditions of 5% carbon dioxide/air for 48 h until exposure to < 1% oxygen conditions, 8% oxygen conditions or standard conditions, while the other half of the cells were exposed to < 1% oxygen, 8%, or standard conditions for 24 h.
Figure 2A-B. DNA methylation patterns at each differentially methylated CpG and its nearby sites. Examples illustrated are (A) CD59 and (B) GRAMD3. Although only the CpG at the site of interest met the criteria for a statistically significant methylation difference (p < 0.05 and Δβ > 0.1), methylation differences are also observed at the nearby sites.
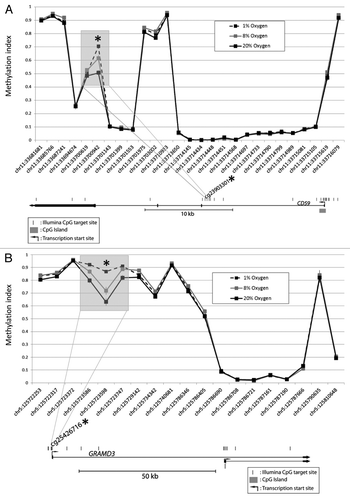
Figure 2C-D. DNA methylation patterns at each differentially methylated CpG and its nearby sites. Examples illustrated are (C) CFB and (D) ZNF217. Although only the CpG at the site of interest met the criteria for a statistically significant methylation difference (p < 0.05 and Δβ > 0.1), methylation differences are also observed at the nearby sites.
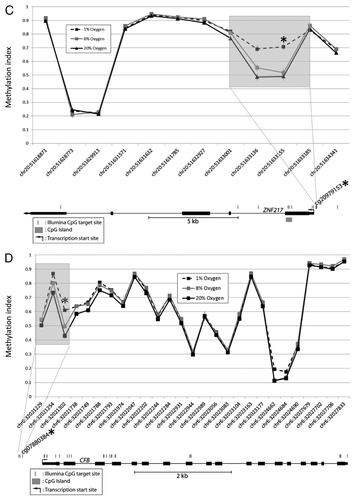
Figure 3. Genomic characteristics of differentially methylated regions. The distribution of CpG loci that were targeted by the methylation array (Array targeted), cytotrophoblast-specifically methylated (Cyto-specific) or hypoxia-specifically methylated (Hypoxia-specific) are shown with respect to different genomic characteristics. RE: repetitive element, *p < 0.0005.
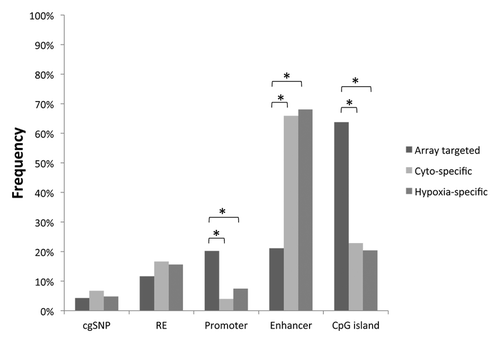
Table 1. Functions of differentially methylated loci between 8% and 20% vs. low (< 1%) oxygen levels
Table 2. Functions of genes associated in common between hypoxia- and cell-phenotype-differentially methylated loci
Figure 4. Motif analysis of hypoxia differentially methylated loci. (A) Position weight matrix description of DNA sequence enriched in hypoxia differentially methylated loci. A consensus DNA sequence of TGACTCA is enriched in 99 out of the 147 loci. (B) The enriched DNA sequence matches the consensus sequence of AP-1 binding region.
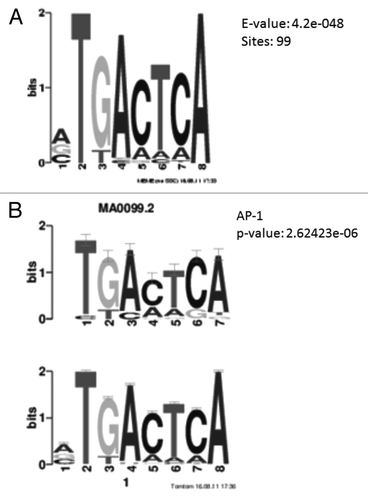
Table 3. Functions enriched for hypoxia-upregulated genes (redundant GO terms related to “ribosome,” “cytosol” and “cell death” were excluded)
Table 4. Functions enriched for hypoxia-downregulated genes (redundant GO terms related to “organelle lumen,” “mitochondrion” and “RNA-binding” were excluded)
Figure 5. Gene expression and DNA methylation correlation of candidate genes. Gene expression as validated by Realtime RT-PCR was shown for (A) JUN, (B) FOS, (C) CFB and (D) CD59. Since the promoters of JUN and FOS are unmethylated, DNA methylation correlation with gene expression was shown only for (E) CFB and (F) CD59.
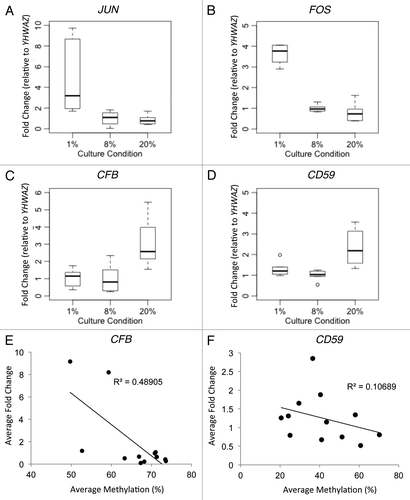
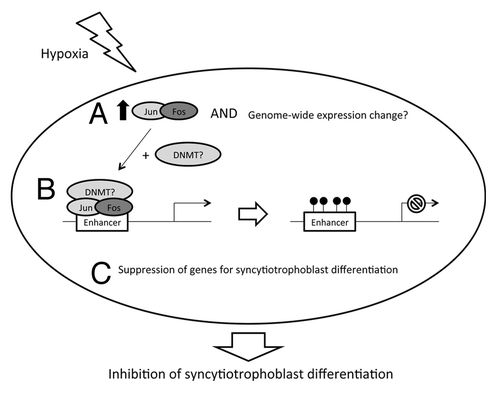
Figure 6. Proposed model of hypoxia effect on cytotrophoblast. Hypoxic exposure may trigger (A) increased expression of JUN and FOS (and perhaps a genome-wide change of gene expression) which encodes Jun and fos to form AP-1 proteins. (B) AP-1 may then recruit DNA methylation machinery (such as DNMTs) for de novo methylation of CpGs at the enhancer regions of various genes. (C) This may cause a suppression of expression for genes that are responsible for syncytiotrophoblast differentiation, which results in depletion of syncytiotrophoblast formation.