Figures & data
Figure 1. Identification of H2AZ lysine methyltransferases. (A) Representation of H2AZWT50-GST. The first 50 amino acids of H2AZ were fused to the N-terminus of GST. (B) Autoradiogram of KMT assays using 3H-SAM, H2AZWT50-GST as a substrate, and the indicated KMTs. The enzymatic reactions were resolved by SDS-PAGE, transferred to PVDF, and either exposed on film (top) or probed with α-GST to show even H2AZ loading. (C) Validation of the potential H2AZ methyltransferases SETD6 and SET7. As in panel B, but full-length recombinant H2AZ-6xHis was used instead of H2AZWT50-GST as the substrate.
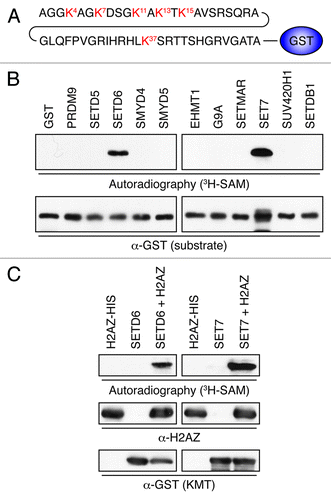
Figure 2. H2AZ is methylated on lysines K4 and K7 in vitro. (A) Representation of the H2AZ mutants. Lysines are highlighted in red. (B) Autoradiogram of the KMT assays using SETD6 (top) or SET7 (bottom) and H2AZ mutants shown in panel A. (C) Flashplate KMT assays using SETD6 and indicated H2AZ-biotin peptides as substrates. Each condition was performed in quadruplicate and the error bars represent the standard error of the mean. The KMT activity is expressed as 3H counts per minute. (D) KMT assays on H2AZWT50-GST were analyzed by immunoblotting using the indicated antibodies. (E) Table summarizing the quantitative MS analysis of SETD6-methylated H2AZ-biotin peptide. (F) Chromatogram of the quantitative MS analysis performed to generate the table in panel E. The molecular weights of mono-methylated K4 and K7 peptides are highlighted (440.29 and 766.49, respectively).
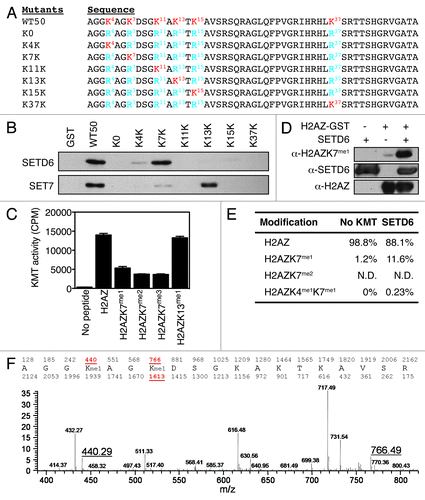
Figure 3. H2AZ is methylated on lysines K4 and K7 in vivo. (A) Protein extracts from U2OS control (LVTHM) or H2AZ knockdown (shH2AZ) cells were analyzed using the indicated methyl-specific antibody. (B) Lentiviruses were used to transduce U2OS cells to express the indicated human SETD6-targeting shRNAs and protein extracts were analyzed by immunoblotting using the indicated antibodies. (C) Immunoblot analysis of protein extracts from shNanog mESC shows loss of Nanog throughout the differentiation time course. (D) Quantitative MS analyses of the H2AZ peptides derived from differentiating mESC induced by Nanog knockdown at the indicated times. *p value of 0.05. (E) Immunoblot analysis of H2AZK7me1 levels in untreated and RA-treated E14 cells. (F) Quantification by MS of total H2AZ, H2AZK7me1, and H2AZK4me1K7me1 from E14 mESC differentiated with RA. * and ** denote p values of < 0.05 and < 0.02, respectively. (G) Quantification by MS of H2AZ acetylation in mESC during shNanog-induced differentiation shown as Abundance (left) and Fold increase (right).
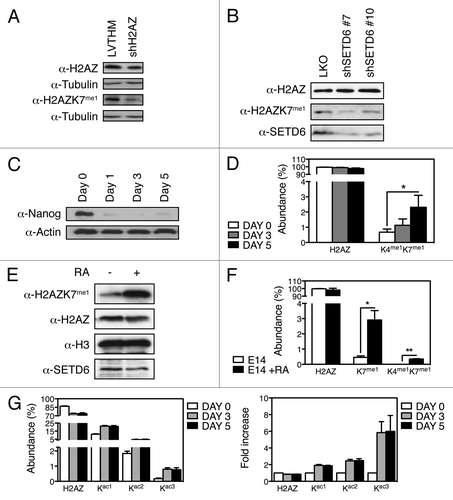
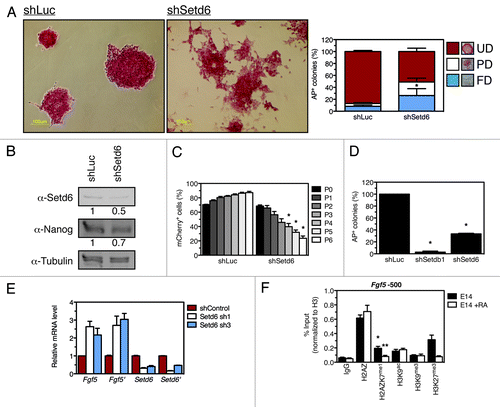
Figure 4. Setd6 maintains mESC self-renewal. (A) E14 mESC were transduced with either shLuc- or shSetd6-expressing lentiviral particles, selected with puromycin, and stained with alkaline phosphatase (AP). AP-stained colonies were scored as fully differentiated (FD), partially differentiated (PD), or undifferentiated (UD) according to the legend. *Denotes a p value of 0.008. (B) Setd6, Nanog, and Tubulin immunoblots from control and shSetd6 mESC. (C) Competition assay in which mCherry-expressing cells (shLuc or shSetd6) were mixed with control cells at a 4:1 ratio, maintained over the course of 6 passages, and monitored for mCherry expression by FACS. *Denotes a p value of < 0.004. (D) Clonogenicity assays in which shLuc, shSetdb1, and shSetd6 cells were plated at clonal density and stained for AP after 10 d. *Denotes a p value of < 0.0002. (E) qPCR validation of Setd6 knockdown and analysis of Fgf5 expression. (F) ChIP analysis of chromatin approximately 500 base pairs upstream of Fgf5 transcriptional start site (TSS) in undifferentiated mESC (E14) or retinoic acid-induced differentiated mESC (E14 + RA). The error bars represent the standard error of the mean calculated from a technical replicate triplicate from a representative biological replicate. *Denotes a p value of < 0.02 between the IgG signal and H2AZK7me1 signal. **Denotes a p value of < 0.05 between the E14 untreated control and the E14 + RA samples.