Figures & data
Figure 1. LECs require LEDGF expression for resistance against UVB-induced damage. (A) UVB radiation caused reduced hLEC viability. Cells were exposed to UVB at different doses (40, 80 and 100 J/m2). After 24h, cell viability was determined by MTS assay. (B) Intracellular ROS were measured at 24h following UVB exposure by replacing the medium with Hank's medium containing 10 μM H2-DCF-DA at Ex485/Em530 nm. (C) LEDGF expression affected cell viability after UVB exposure. MTS assay was performed to examine viability of cells under- or overexpressing LEDGF after UVB exposure (40, 80 and 100 J/m2). When LEDGF was depleted in hLECs by Si-LEDGF, the cells became more vulnerable to UVB-induced damage, while LECs overexpressing LEDGF had resistance against UVB stress. Transfection efficiency was normalized with GFP OD values measured at ex485/em530. Values are mean ± SEM of three independent experiments. Asterisks indicate statistically significant difference (p < 0.001 vs. control). (D) Representative Western analysis for LEDGF protein to show effective silencing of LEDGF in hLECs with specific siRNA strategy. (E) Diagrammatic representation of UVB stress schedule conducted in the study.
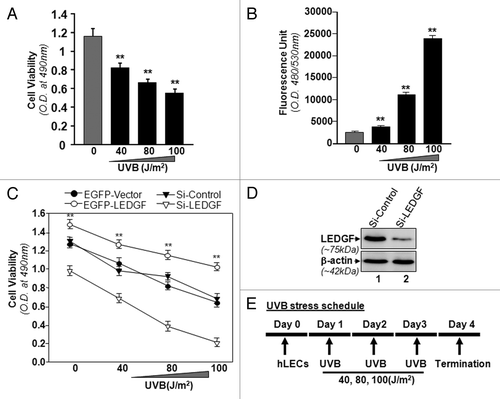
Figure 2. Effect of single or multiple doses of UVB exposure on the expression of LEDGF mRNA. (A) Single exposures to variable doses of UVB differentially enhanced LEDGF expression in LECs. Cultured cells were exposed one time to different doses of UVB radiation as shown. Real-time PCR analysis was performed with mRNA isolated from hLECs, and expression of LEDGF mRNA was normalized with β-actin. Values are mean ± SD of three independent experiments. Asterisks indicate statistically significant difference (p < 0.001 vs. control). (B) Multiple high doses (80 and 100 J/m2) of UVB exposure suppressed the LEDGF mRNA expression. Real-time PCR analysis was performed with mRNA isolated from hLECs after multiple exposures to UVB (three times at 24h intervals) . Values are mean ± SD of three independent experiments. Asterisks indicate statistically significant difference (p < 0.001 vs. control). (C) LECs exposed to multiple high doses (80 and 100 J/m2) of UVB radiation displayed reduced LEDGF protein expression. Cells were either unexposed or exposed to UVB radiation three times at variable doses at 24h intervals. After 96h, nuclear extract was isolated, resolved onto SDS-GEL and immunoblotted using antibody specific to LEDGF. The membrane was striped or restriped and reprobed with β-actin antibody for internal/loading assessment.
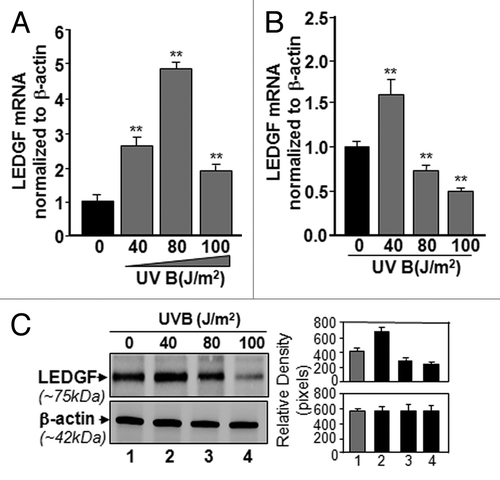
Figure 3. TSA, an HDAC inhibitor, was able to relieve repressed LEDGF promoter-driven CAT activity in LECs or LECs facing multiple UVB exposures, but 5-AzaC was not. Promoter activity assay was performed by co-transfecting LECs with LEDGF-CAT reporter plasmid (-170/+35bp) or empty CAT vector along with pSEAP vector. Transfectants were exposed to variable doses of UVB radiation as shown. Cells were pretreated as follows: (A) untreated control; (B) TSA treated; (C) 5-AzaC treated, followed by UVB. After 48h cell lysate was prepared to measure CAT activity. Transfection efficiencies were normalized with protein concentration and SEAP values (OD; ex/em, 360/449). Values are mean ± SD of three independent experiments. Asterisks indicate statistically significant difference (p < 0.001 vs. control).
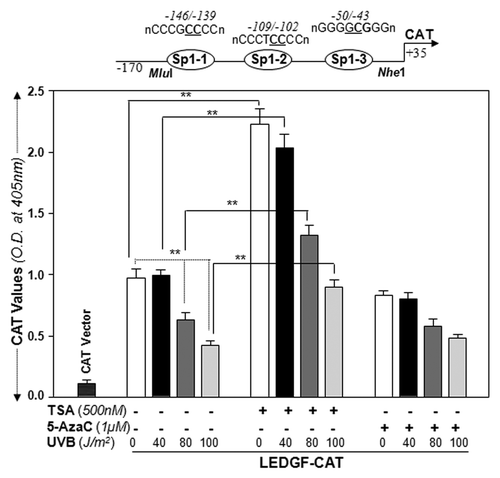
Figure 4. LECs exposed to UVB displayed enhanced expression of DNA methyl transferase DNMT3b, but could not methylate LEDGF promoter. (A) Nuclear extract from UVB exposed hLECs were resolved on 4–20% SDS gel and immunoblotted using anti-DNA methyl transferases as shown. The same membranes were used to re-probe the antibodies following restriping. Protein band appearing by β-actin antibody served as internal/loading control. Right panel shows relative densitometry of protein bands. (B) MSP analysis disclosed that status of LEDGF promoter (-175/+27) containing Sp1 sites within the CpG island was not altered. Top panel shows the diagrammatic representation of the LEDGF promoter, predicted CpG island and regions where the PCR primers bind. Lower panel shows the representative gel image from MSP analysis of genomic DNA isolated from LECs exposed to variable doses of UVB-exposed or unexposed to UVB.
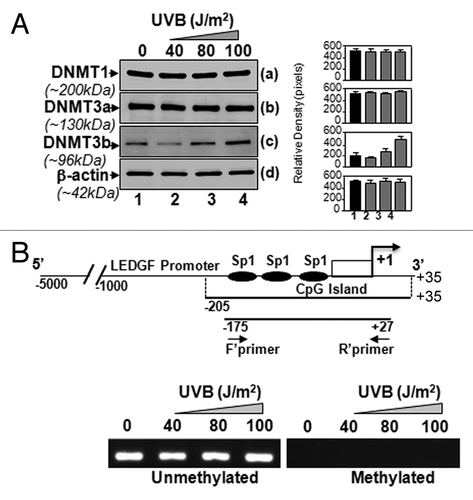
Figure 5. UVB-induced differential expression of posttranslationally modified acetyl-H3 and H2B was associated with decreased total HAT activity. (A) UVB-exposed and unexposed LECs were harvested and processed for nuclear protein extraction. Extract was resolved through 4–20% SDS-PAGE and immunoblotted with anti-acetyl histone or anti-histone antibodies as shown. (B) Total HAT activity was measured after multiple UVB exposures following the company’s protocol (Epigentek), and data were compared with results from untreated samples. The data represent mean ± SEM of three independent experiments. Asterisks indicate statistically significant difference (p < 0.001 vs. corresponding control).
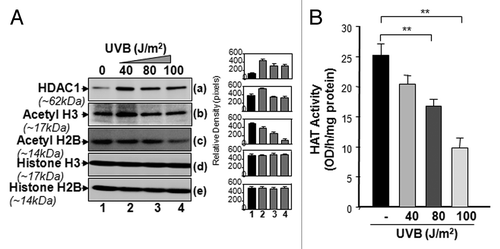
Figure 6. ChIP analyses revealing reduced binding of Sp1 and the status of histone acetylation associated with LEDGF promoter. Left panel shows Sp1 binding to its responsive cis-elements (B), association and recruitment of site-specifically modified H3 (C), H2B (D), and HDAC1(E) to the Sp1 binding sites within CpG island of LEDGF gene promoter in LECs exposed or unexposed to UVB radiation. Right panel represents control; DNA selected beyond Sp1 binding sites is shown. ChIP assay was performed. Also, amplification of chromatin extract before precipitation is shown as control (input, A)
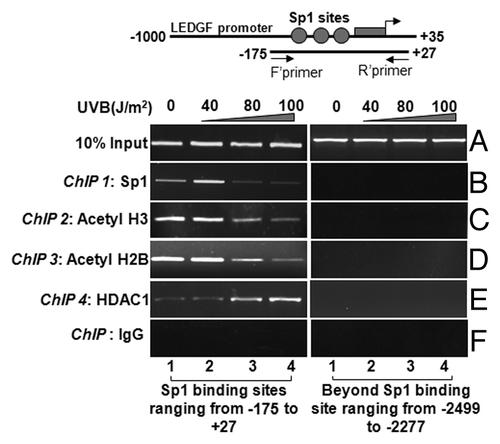
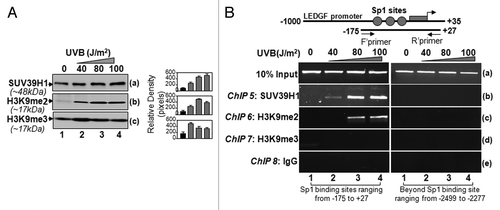
Figure 7. Sp1 binding sites within CpG island in LEDGF promoter were enriched with SUV39H1 and dimethylated histones in LECs. (A) Western analysis of SUV39H1, H3K9me2, and H3K9me3 in the extract isolated from LECs exposed to UVB radiation. Right panel shows the relative densitometry of protein bands. (B) ChIP analyses revealed recruitment of histone methyltransferase SUV39H1 and enrichment of H3K9me2 at CpG island containing Sp1 sites. Chromatin immunoprecipitation analysis was performed using chromatin extract from LECs exposed to multiple doses of UVB radiation as shown in the scheme () with specific antibodies, as indicated. Top panel shows LEDGF promoter and location of PCR primers designed for the study. DNA fragments present in the immunoprecipitates were amplified with primers that specifically recognize a fragment of the LEDGF promoter containing Sp1 binding sites (-175 to +27). As a negative control, primers designed beyond the Sp1 sites (-2499 to -2277) or a mock ChIP with control IgG was used. PCR products were separated on 2.5% agarose gels and stained with ethidium bromide.