Figures & data
Table 1. PCR primers forward (F), reverse (R) and sequencing primers (S) used for the FOLR1 gene Pyrosequencing methylation assays
Table 2. PCR primers forward (F), reverse (R) and sequencing primers (S) used for the PCFT gene Pyrosequencing methylation assays
Table 3. PCR primers forward (F), reverse (R) and sequencing primers (S) used for the RFC1gene Pyrosequencing methylation assays
Figure 1. Methylated fraction in the FOLR1 gene in normal placenta (n = 4), leukocytes from subjects with low tHcy (c = 5–10 µmol/L, n = 24–25), and leukocytes from subjects with high tHcy (c = 20–113 µmol/L, n = 23–25). The error bars show ± 1 SD.
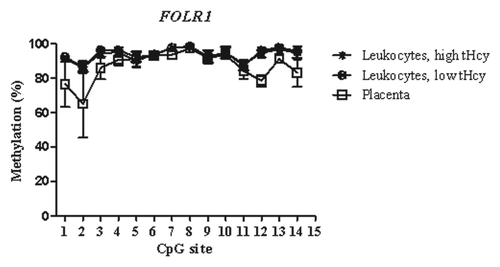
Figure 3. Methylated fraction in the RFC1 gene in normal placenta (n = 4), leukocytes from subjects with low tHcy (c = 5–10 µmol/L, n = 22–24), and leukocytes from subjects with high tHcy (c = 20–113 µmol/L, n = 21–24). The error bars show ± 1 SD. The upper panel shows CpG sites 1–24 and the lower panel shows CpG sites 25–55.
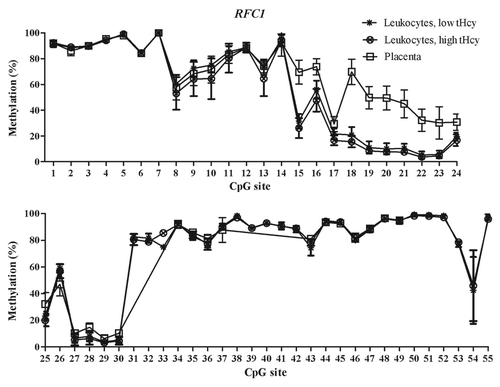
Table 4. mRNA expression of FOLR1, PCFT and RFC1 in full-term placental tissue and whole blood leukocytes
Figure 2. Methylated fraction in the PCFT gene in normal placenta (n = 4), leukocytes from subjects with low tHcy (c = 5–10 µmol/L, n = 10–25), and leukocytes from subjects with high tHcy (c = 20–113 µmol/L, n = 11–25). The error bars show ± 1 SD. The upper panel shows CpG sites 1–30 and the lower panel shows CpG sites 31–52.
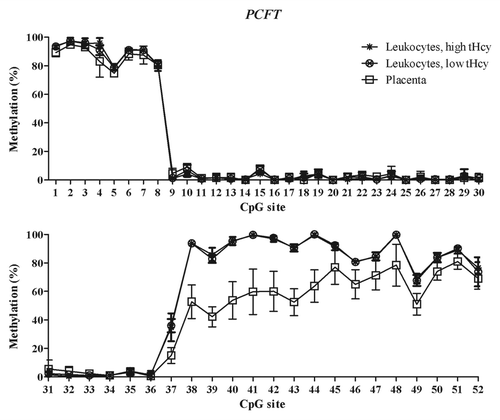
Table 5. Mean methylated fraction of CpG sites in the RFC1 gene in subjects with low or high plasma tHcy concentration
Figure 4. Methylated fraction in the FOLR1, PCFT and RFC1 gene in placental tissue from healthy (n = 39–48) and NTD (n = 66–75) subjects. The error bars show 1 ± SD.
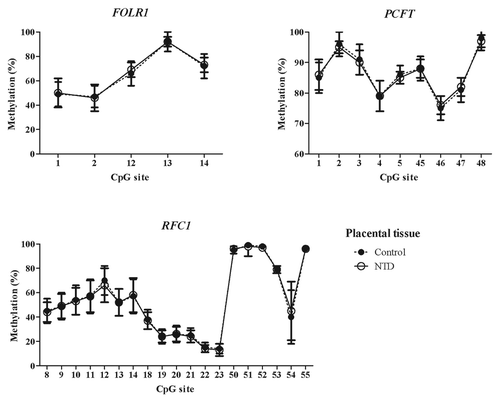
Table 6.RFC1 80G > A genotype prevalence and allele frequencies in subjects with low and high tHcy and normal and NTD subjects
Table 7. Methylated fraction of CpG sites in the RFC1 gene in blood leukocytes form subjects with low and high tHcy according to genotype
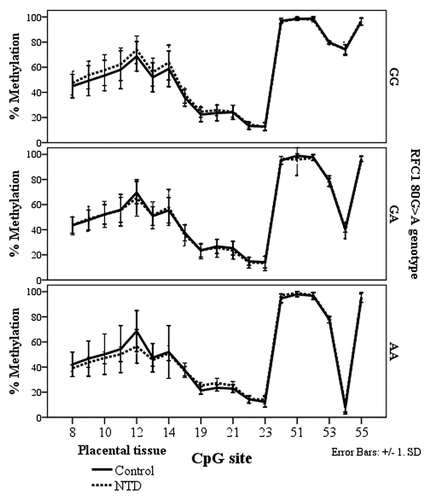
Figure 5. Methylated fraction in the RFC1 gene according to RFC1 80G > A genotype in placentas, stratified by groups of healthy controls (n = 38) and NTD (n = 68). The error bars show ± SD.
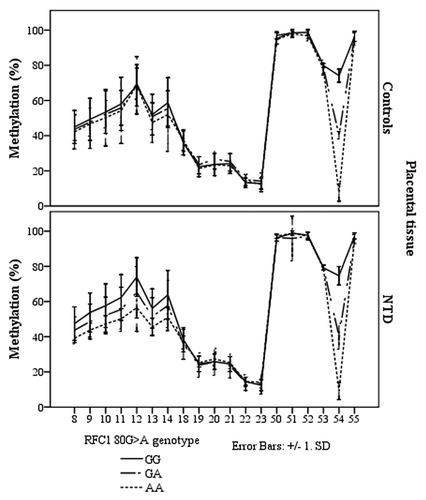
Figure 6. Methylated fraction in the RFC1 gene in placentas of normal control births (n = 38) and NTD births (n = 68) stratified by RFC1 80G > A genotype.