Figures & data
Figure 1. HLCS facilitates methylation events in the epigenome through physical interactions with DNA methyltransferase 1 (DNMT1), methyl CpG binding protein 2 (MeCP2) and eukaryotic histone methyltransferase (EHMT-1).
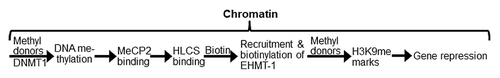
Figure 2. HLCS interacts with both MeCP2 and DNMT1 in vivo, judged by co-immunoprecipitation assay. Myc-HLCS and HA-DNMT1 were overexpressed in HEK293 cells and protein extracts were immunoprecipitated using anti-Myc (A) and anti-HA (B). Proteins were resolved by electrophoresis and probed with anti-DNMT1 (A) and anti-HLCS ( B).Panels C and D are similar to A and B. But HA-MeCP2 was substituted for HA-DNMT1 in overexpression experiments and anti-MeCP2 (C) and anti-Myc (D) were substituted for anti-DNMT1 and anti-HLCS, respectively. Transfections with empty vectors were used as negative controls (second lane in each gel), and whole cell lysates were used as input controls (third and fourth lane in each gel). Gels were electronically re-arranged to facilitate comparisons.
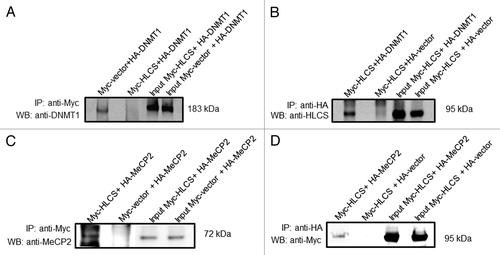
Figure 3. HLCS interacts with both MeCP2 and DNMT1 in vitro, judged by limited proteolysis assay. (A) Purified recombinant GST-HLCS protected His-DNMT1 from trypsin digestion (left panel) compared with DNMT1 that was pre-incubated with GST (right panel). (B) Purified recombinant GST-HLCS protected His-MeCP2 from trypsin digestion (left panel) compared with MeCP2 that was pre-incubated with GST (right panel).
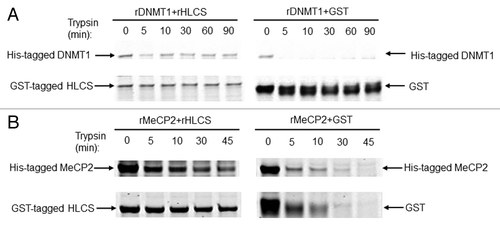
Figure 4. Transfection with plasmid FLAG/Myc-HLCS produced a stable overexpression of HLCS compared with non-transfected controls. The Myc-tagged HLCS in cell extracts was probed with anti-HLCS and anti-Myc. GAPDH was probed as loading control.
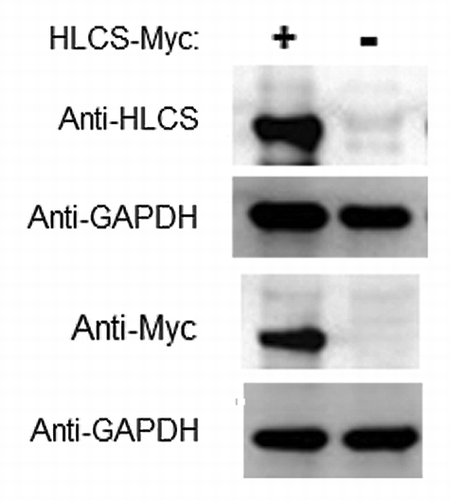
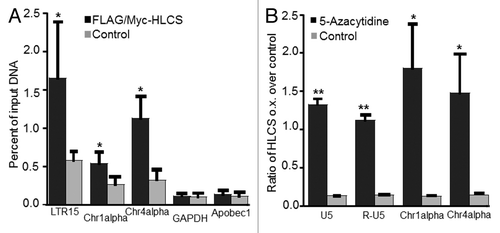
Figure 5. The effects of HLCS on LTR repression depend on DNA methylation. (A) The enrichment of H3K9me3 marks in loci coding for LTR15, Chr1alpha, Chr4alpha, GAPDH and Apobec1 was quantified by chromatin immunoprecipitation assay/qRT-PCR in HEK293 HLCS overexpression cells and control cells (H = 6.9018, 1 d.f., *p-value = 0.0086 HLCS overexpression vs. control; H = 4.8402, 1 d.f., *p-value = 0.0278 for Chr1alpha; H = 6.8598, 1 d.f., p-value = 0.0088 for Chr4alpha; H = 0.1756, 1 d.f., p-value = 0.6752 for GAPDH; H = 0.7245, 1 d.f., p-value = 0.3947 for Apobec1). (B) The transcription of LTRs, Chr1alpha and Chr4alpha was repressed in HLCS overexpression HEK293 cells compared with controls. Treatment of 5-azacytidine (1 µM) abrogated the effects of HLCS on repeat repression (**p-value < 0.0001, *p-value < 0.05, 5-azacytidine treatment vs. control).