Figures & data
Figure 1. Late passage WI-38 fibroblasts underwent telomere shortening and display a senescent phenotype. (A) TRF analysis on genomic DNA isolated from early (E) passage and late (L) passage cells. DNA was digested with RsaI and AluI and reduction of the average telomere length is seen by the shift in the smear of the undigested telomeric DNA. (B) Normalized relative quantification of telomeric copy number in E and L passage cells. Telomeric copy number was quantified by qPCR, relative to the 36B4 single copy locus and normalized to E passage cells. Error bars indicate SEM of a triplicate measurement. (C) Western blot analysis in of p16 and actin expression in E and L passage cells followed by (D) relative quantification, showed increased expression of p16 in L passage cells. Error bars indicate stdev of two biological replicates, asterisks indicate a p-value < 0.05 based on a student’s t-test.
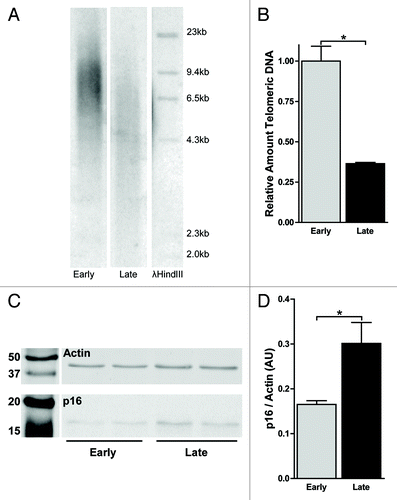
Figure 2. Decreased levels of markers of constitutive heterochromatin at subtelomeres upon senescence. (A) Location of the generated primer pairs along the subtelomeres of chromosome 7q and 11q. (B) Relative quantification using ChIP-qPCR of H3K9me3 along the subtelomeres of 7q and (C) 11q shows a decrease at all loci upon senescence. H3K9me3 levels are corrected for IgG background and normalized to the relative amount of H3: (CtIgG - CtH3K9me3)/(CtIgG - CtH3). Error bars indicate SEM of at least duplicate qPCR measurements, asterisks indicate a p-value < 0.05 based on a student’s t-test. (D) EpiTYPER based quantification of CpG methylation along the subtelomere of 7q and (E) 11q showed a decreased DNA methylation at all sites measured. The mean methylation of multiple CpGs within the indicated probes is displayed. (F) Combined mean methylation levels of all subtelomeric probes at 7q and 11q showed a decreased DNA methylation along both subtelomeres. DNA methylation was quantified in triplicate, p-values are indicated and * indicates a p-value < 0.05 based on UNIANOVA analysis.
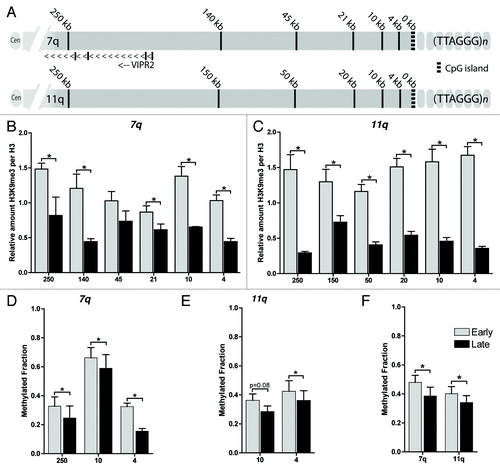
Figure 3. Relative quantification of subtelomeric H4K16ac, H3K27me3 and H3K36me3 upon senescence. Relative quantification using ChIP-qPCR of H4K16ac (A, B), H3K27me3 (C, D) and H3K36me3 (E, F) at increasing distance from the telomere at the subtelomeres of 7q (A, C, E) and 11q (B, D, F). H4K16ac is decreased at all loci and H3K36me3 shows an increase at both subtelomeres. H3K27me3 shows a more varied pattern; an increase is observed at 7 out of 12 loci. All values are corrected for IgG background and normalized to the relative amount of H3 [(CtIgG - Ctmodification)/(CtIgG - CtH3)], error bars indicate SEM of duplicate qPCR measurements, asterisks indicate a p-value < 0.05 based on a student’s t-test.
![Figure 3. Relative quantification of subtelomeric H4K16ac, H3K27me3 and H3K36me3 upon senescence. Relative quantification using ChIP-qPCR of H4K16ac (A, B), H3K27me3 (C, D) and H3K36me3 (E, F) at increasing distance from the telomere at the subtelomeres of 7q (A, C, E) and 11q (B, D, F). H4K16ac is decreased at all loci and H3K36me3 shows an increase at both subtelomeres. H3K27me3 shows a more varied pattern; an increase is observed at 7 out of 12 loci. All values are corrected for IgG background and normalized to the relative amount of H3 [(CtIgG - Ctmodification)/(CtIgG - CtH3)], error bars indicate SEM of duplicate qPCR measurements, asterisks indicate a p-value < 0.05 based on a student’s t-test.](/cms/asset/a6cd6567-aaac-4327-b95c-bc8082e49d1d/kepi_a_10924450_f0003.gif)
Figure 4. TERRA promoter analysis reflects transcriptional activity at 7q and 11q irrespective of senescence. (A) RT-PCR analysis of 7q, 11q and Xq specific TERRA transcripts allowed detection of 11q, but not 7q specific transcripts. Xq serves as an internal control. –RT: cDNA synthesis in the absence of reverse transcriptase to control for DNA background. gDNA: positive control for the PCR on a WI-38 genomic DNA extract. (B) Epityper based quantification of TERRA promoter CpG methylation at 7q and 11q showed higher levels of methylation at 7q pTERRA compared with 11q pTERRA. The mean methylation of multiple CpGs within the indicated probes is displayed. Upon senescence TERRA promoter methylation at both 7q and 11q does not change. Indicated p-values (* < 0.05) were obtained using an UNIANOVA analysis of triplicate measurements. (C) Relative quantification by ChIP-qPCR of H3K4me3, marking active promoters, showed higher levels at 11q pTERRA compared with 7q pTERRA, irrespective of senescence. All values are corrected for IgG background and are normalized to the relative amount of H3 (CtIgG - CtH3K4me3)/(CtIgG - CtH3). Error bars indicate SEM of duplicate qPCR measurements, asterisks indicate a p-value < 0.05 based on a student’s t-test.
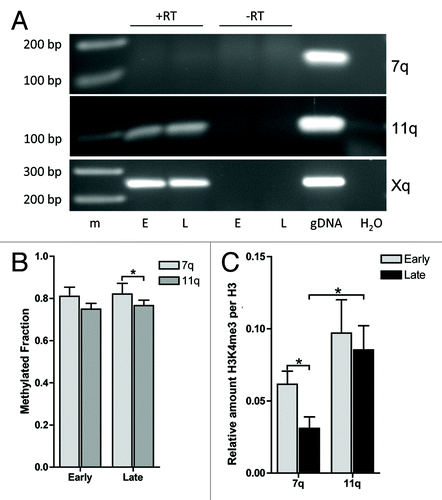
Figure 5. Distinct chromatin remodeling at TERRA promoters of 7q and 11q upon senescence. TERRA promoter analysis showed distinct regulation of (A) H3K9me3, (B) H4K16ac, (C) H3K27me3 and (D) H3K36me3 between 7q and 11q. The changes at the 7q TERRA promoter reflect the more proximal subtelomere, whereas the 11q TERRA promoter regulation is distinct from the more proximal subtelomere. All values are corrected for IgG background and are normalized to the relative amount of H3 [(CtIgG -Ctmodification)/(CtIgG - CtH3)]. Error bars indicate SEM of duplicate qPCR measurements, asterisks indicate a p-value < 0.05 based on a student’s t-test.
![Figure 5. Distinct chromatin remodeling at TERRA promoters of 7q and 11q upon senescence. TERRA promoter analysis showed distinct regulation of (A) H3K9me3, (B) H4K16ac, (C) H3K27me3 and (D) H3K36me3 between 7q and 11q. The changes at the 7q TERRA promoter reflect the more proximal subtelomere, whereas the 11q TERRA promoter regulation is distinct from the more proximal subtelomere. All values are corrected for IgG background and are normalized to the relative amount of H3 [(CtIgG -Ctmodification)/(CtIgG - CtH3)]. Error bars indicate SEM of duplicate qPCR measurements, asterisks indicate a p-value < 0.05 based on a student’s t-test.](/cms/asset/c2433950-e8fc-493f-a8f4-60ce3e239dff/kepi_a_10924450_f0005.gif)