Figures & data
Figure 1. DMC, DVCs and DMVCs. (A) Density histograms of DMCs (differentially methylated CpG sites). These P-values were derived from linear regression based on the Limma package comparing differences in means between lean and obese subjects. (B) Density histograms of DVCs (differentially variable CpG sites). These P-values were derived from Bartlett’s test comparing differences in variances between lean and obese. (C) Venn diagram illustrating DMCs, DVCs and DMVCs (differentially methylated and variable CpG sites). The overlapping DMVCs were significantly enriched with P.Fisher < 2.2E-16 and P.permutation (1000 times) < 0.001. (D) Top ranked DMC cg08339189. Y-axis shows the β value, x-axis the sample. Phenotypes were indicated as lean (black, n = 48) and obese (red, n = 48). The dashed lines show the mean levels in lean (0.15) and obese (0.18) separately. (E) Top ranked DVC cg24570070. The mean levels were not significantly different (0.88 in obese vs. 0.91 in lean). However, obese cases showed a large methylation variance. (F) Top ranked DMVC cg00033915. The mean levels (dashed line) were significantly different between the two groups (0.92 in obese vs. 0.93 in lean, raw p = 1.1E-5 and FDR = 4.2E-3). Furthermore, the obese group also showed significantly larger variance.
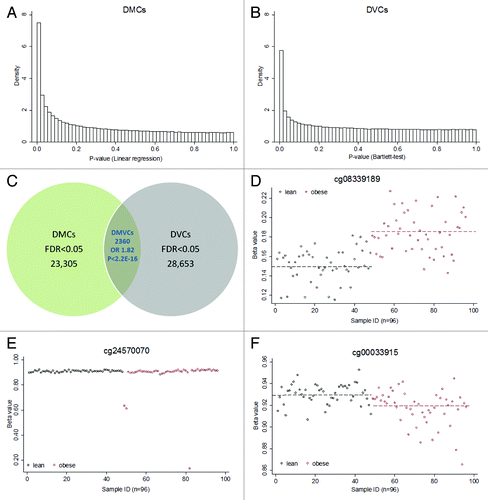
Figure 2. Distributions of DMCs, DVCs and DMVCs. (A) Distributions across average methylation levels. The Y-axis represents the density, x-axis the average methylation β value. The orange line presented the kernel density curve of overall CpG sites across β 0–1, the green line presented DMCs, the gray line DVCs and the blue line DMVCs. The percentages of each of these four types of CpG sites with betas below 0.2 or above 0.8 are also listed in the plot. (B) Distribution across genomic locations. Chi-square test found significant difference among their distributions across the genome with P-value < 2.2E-16. TSS200, CpG sites within 200bp from the transcription starting site (TSS); TSS1500, CpG sites within 200–1500bp from the transcription starting site (TSS); body, gene body.
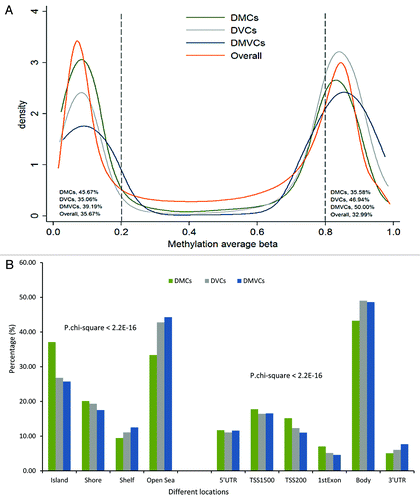
Figure 3. Increased methylation variance in obese. (A) Scatter plot of mean methylation difference (x-axis) against methylation variance ratio (y-axis) comparing lean and obese. The percentage of DMCs, DVCs and DMVCs within each of the four quadrants was also listed. (B) Scatter plot of mean methylation difference (x-axis) against methylation variance ratio (y-axis) comparing lean and obese limited to the CpG sites on the illumina 27K chip. The percentage of DMCs, DVCs and DMVCs within each of the four quadrants was also listed.
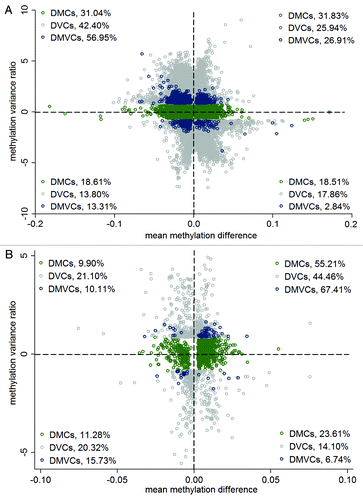
Figure 4. Predictive ability of DMCs and DVCs. (A) Predictive ability of DMCs. The receiver operating characteristic (ROC) analysis showed the area under curve (AUC) and its 95% confidence interval (CI) in a randomly split testing set (24 obese vs. 24 lean) from the whole data set. (B) Predictive ability of DVCs. AUC and 95%CI was presented in a randomly split testing set (24 obese vs. 24 lean) from the whole data set.
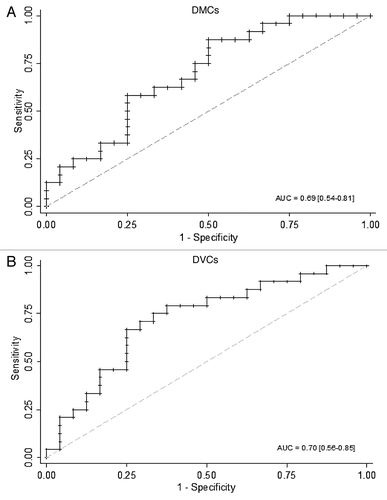
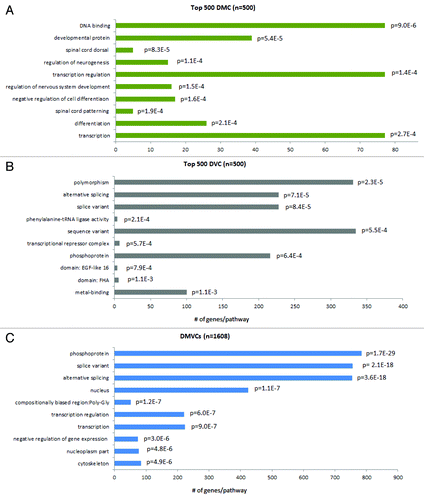
Figure 5. Gene ontology enrichment analysis of DMCs, DVCs and DMVCs. Gene ontology analysis was performed using DAVID (http://david.abcc.ncifcrf.gov). The human genome was used as background. The top 500 DMCs (A), the top 500 DVCs (B) and all DMVCs (n = 1608) (C) were selected for analysis. The top ten enriched pathways are listed here together with their enrichment P-values, which are derived from a modified Fisher’s exact test.