Figures & data
Figure 1. Characterization of the antibodies against mono-, di- and tri-methyl lysine. (A) Dot blot analysis of the pan anti-methyl lysine antibodies. Peptides with un- (Kun), mono- (Kme1), di- (Kme2) and tri- (Kme3) methyl lysine were blotted onto nitrocellulose membranes and probed with the antibodies, respectively. (B) Immunoprecipitation (IP) analysis of lysates prepared from HeLa cells using the antibodies. Lanes A, B and C are input, IP eluates with normal rabbit IgG (negative control) and IP eluates with anti-methyl lysine antibodies, respectively. The concentration of SDS-PAGE was 15%. (C) Lysine methylation profiles in 13 human cell lines. The protein lysates of 13 cell lines were separated by 12.5% SDS-PAGE, transferred to PVDF membranes and probed with the antibodies, respectively.
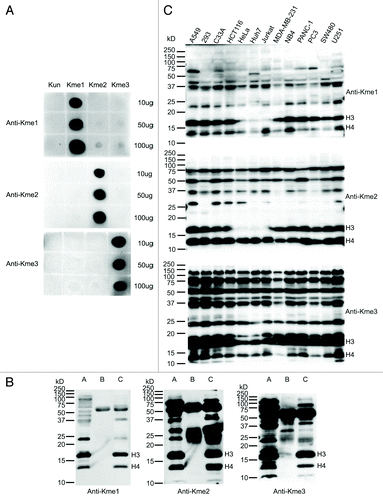
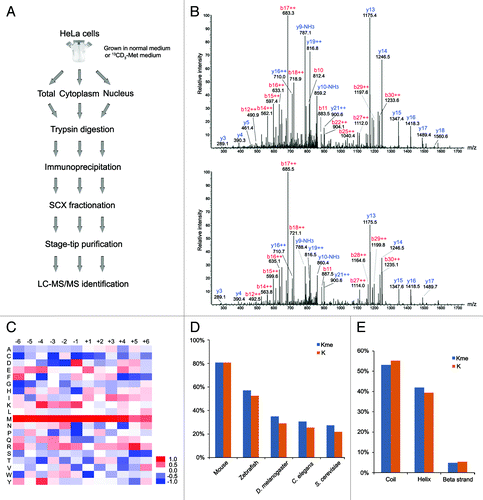
Figure 2. Illustration of the study strategy and characterization of the methylation sites. (A) Illustration of the study strategy. HeLa cells cultured in normal or 13CD3-methionine medium were lysed, or sub-cellularly fractionated to separate cytoplasm and nucleus, and their protein lysates were digested by trypsin. The methylated peptides were enriched through immunoprecipitation and underwent strong cation-exchange (SCX) separation. Each fraction was desalted and subjected to mass spectrometry. (B) MS/MS spectra of the methylated peptide, TKme1AAAAAAAAAPAAAATAPTTAATTAATAAQ. The upper and lower spectra come from normal and heavy methyl-labeled samples, respectively. (C) Heat map of relative distribution of amino acids around the methylated lysine sites. Amino acids from -6 to +6 positions adjacent to the methylated lysine sites and all lysine residues were extracted, respectively. At each position, the frequency of amino acid around methylated lysine sites was divided by that around all lysine residues, and the ratios were visually represented after logarithmic transformation (log2). Color scale reflects the degree of enrichment (red) or depletion (blue). (D) Evolutionary conservation of the methylated lysine sites and all lysines throughout the eukaryotic species. “Kme” and “K” denote methylated lysines and all lysines, respectively. (E) Distribution of methylated lysines and all lysines in protein secondary structure. “Kme” and “K” denote methylated lysines and all lysines, respectively.