Figures & data
Table 1. Summary of the P35S epilallelic variants reported in different systems and their molecular characteristics.
Figure 1. Schematic outline of T-DNA insertions in locus 1, locus 2 and locus 271. Locus 1 contains two copies of T-DNA that are arranged as an IR about the right border. Promoters are in hatched squares; P35S, CaMV 35S promoter; P19S, CAMV 19S promoter; nptII, neomycin phosphotransferase II gene; RIN, nitrate reductase gene in an antisense orientation. M bar indicates region analyzed by bisulfite genomic sequencing. P (promoter) and E (3?end) bars correspond to PCR fragments amplified after ChIP. RB, LB right and left border, respectively.
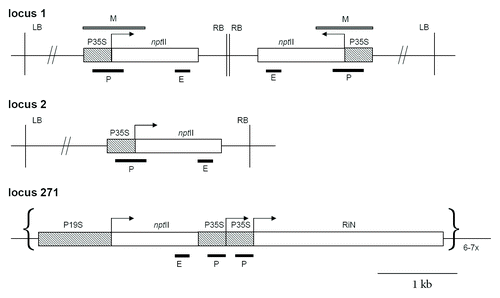
Figure 2. Schematic outline of P35S epialleles generation. (A) Epialleles generated spontaneously in callus culture. (B) Epialleles induced by siRNAs in 271 Locus 1 hybrids. S1, S2 represent plants of first or second generation after silencer locus segregation respectively. ?x NT,? cross to a non-transgenic tobacco.
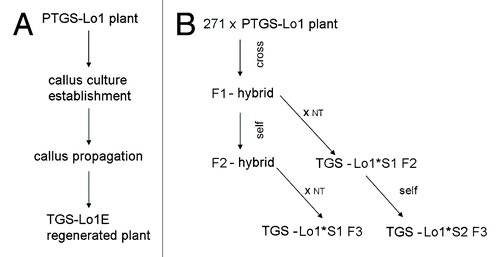
Figure 3. Expression analysis of the nptII reporter genes in parental plants and derived epiallelic variants. The nptII transcripts were analyzed by quantitative RT-PCR. (A) epialleles generated by callusogenesis. Results include data from two independent experiments (3 technical replicates); (B) transcription level of epialleles generated by siRNA signals. Values are expressed as percentages of the nptII levels in a non-silenced HeLo2 line.
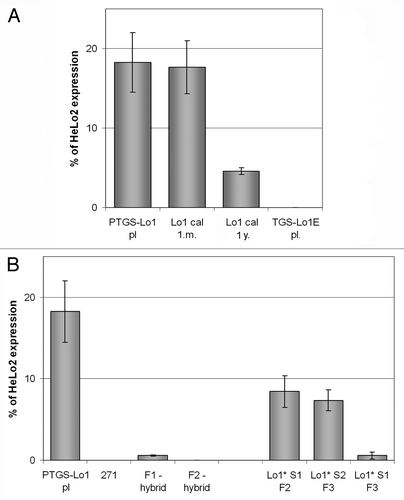
Figure 4. Detailed bisulfite methylation analysis of epiallelic variants. (A) Transgene subregions subjected to bisulfite sequencing. (B) Column graphs showing average methylation levels from 5?10 clones. Error bars represent clones with maximum and minimum level of DNA methylation. (C) Distribution of mC along the sequenced fragment. Individual vertical columns represent the average methylation at particular position. Positions of the as-1 regulatory element and Sau96I restriction sites (asterisks) are indicated.
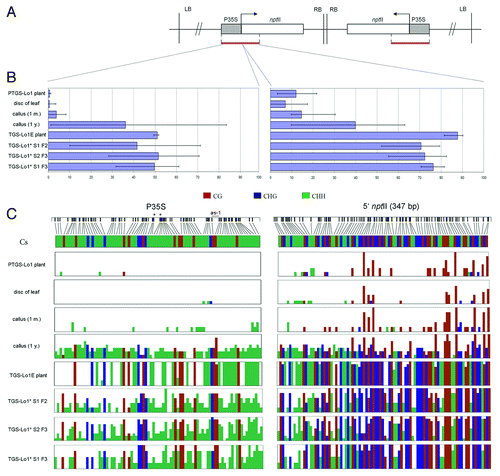
Figure 5. Histone modification patterns along the transgenes. (A) Schematic outline of subregions analyzed by ChIP: P (promoter) and E (3? end of nptII). The P subregion in locus 271 was shorter (upper line) than in loci 1 and 2 (bottom line). (B) Electrophoretic profiles of PCR products obtained by amplification of immunoprecipitated DNA. Results of a repeated experiment performed on an independent callus culture are shown in Figure S3.
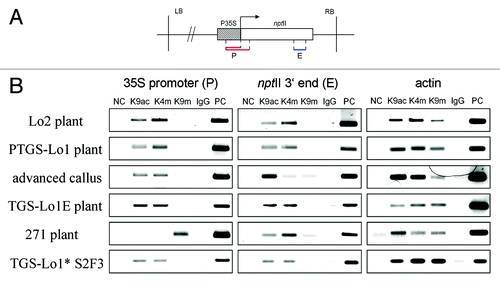
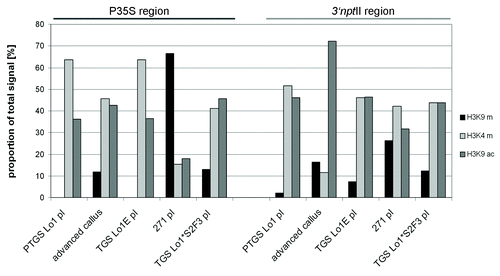
Figure 6. Semiquantitative evaluation of histone modification patterns along the transgenes. Fluorescent signals in gels () were counted and the levels were expressed as percentages of total signal (a sum of H3K9me2, H3K4me3 and H3K9ac).