Figures & data
Figure 1. Reduced C2ORF40 mRNA expression was a significant prognostic factor for disease-free and metastasis-free survival. C2ORF40 expression assessed by Affymetrix microarray in human breast cancers and normal tissues in data set 1 to 3 (Table S1) was shown in panel (A) to (C). The significant association between C2ORF40 mRNA level and disease-free survival was analyzed in four independent cohorts of breast cancer patients (Data set 4 to 7 in Table S1) (D) to (G). The patients from each cohort were divided into groups with high (top one-third), moderate (middle one-third) and low (bottom one-third) level of C2ORF40 expression. Panel (D) to (G) show the Kaplan-Meier survival curves for disease-free survival in the four data sets respectively. Panels (H) and (I) show the Kaplan-Meier survival curves for metastasis-free survival in Data set 4 (NKI) and Data set 7 (GSE6532) respectively. C2ORF40 mRNA is measured as log2 (probe intensities) as in the microarray. The P-values shown were obtained from Mann-Whitney U (A and B), Kruskal-Wallis (C) or long-rank tests (D to I).
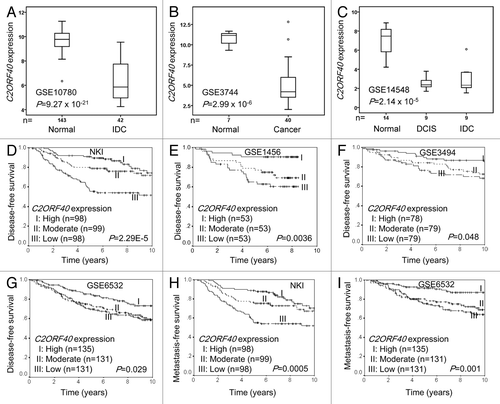
Figure 2. Loss of C2ORF40 expression due to promoter hypermethylation. (A) Schematic map of the 5′-CpG islands located in the C2ORF40 promoter region around the transcription start site (ATG). CpG dinucleotides are depicted. PCR primers used for sodium bisulfite sequencing (SF1, SF2, SR1 and SR2) and for detecting methylation (MF and MR) and non-methylation (UF and UR) are indicated as arrows. The sequences of these primers are listed in Materials and Methods. (B) The representative sequence traces show the methylation of C2ORF40 promoter. (C) Analysis of C2ORF40 promoter methylation using MSPCR. U indicates unmethylated, M indicates methylated. (D) The relationship between C2ORF40 expression and its promoter methylation in a panel of human breast cancer cell lines. (E) qRT-PCR shows C2ORF40 expression is lower in promoter hypermethylated MDAMB231and BT549 cells than in unmethylated SKBR3, HCC70 cell lines and 5-aza-dC treatment restored C2ORF40 expression in MDAMB231 and BT549 cell lines. (F) The relationship between C2ORF40 expression and its promoter methylation in human primary breast cancers. Each experiment in (B), (C) and (E) was repeated at least three times. Data were presented as boxplot based on C2ORF40 promoter methylation status in (D) and (F) and mean ± SD in (E). The P-values shown in (D) and (F) were obtained from nonparametric test. * p < 0.05 and ** p < 0.01 in (E) obtained from Student’s t-test as compared with control groups.
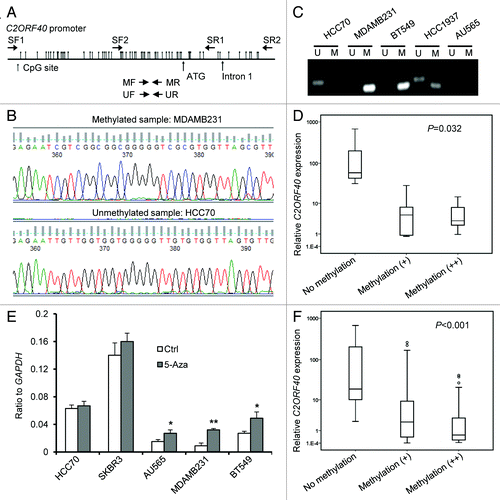
Table 1.C2ORF40 promoter methylation status in human breast cancer cell lines
Figure 3. Ectopic expression of C2ORF40 inhibits breast cancer cell growth and migration-invasion. (A) C2ORF40 is stably overexpressed in retroviral transduced BT549 and MDAMB231 cells as indicated by RT-PCR (upper panel) and western blotting (lower panel). Overexpression of C2ORF40 inhibits BT549 and MDAMB231 cell proliferation assessed by MTT method (B, C) and colony formation assay (D, E). (B, D) for BT549 and (C, E) for MDAMB231 cells, respectively. Representative photographs show ectopic expression of C2ORF40 inhibits migration of BT549 (F) and MDAMB231 (G) cells in a scratch-wound healing model on cultured cells. Ectopic expression of C2ORF40 in BT549 (H) and MDAMB231 (I) cells decreases cancer cell invasive abilities using Matrigel assay. All experiments were repeated at least three times in two independently retrovirally transduced cell lines. Data were presented as mean ± SD, * p < 0.05 and ** p < 0.01 obtained from Student’s t-test as compared with control groups. Ctrl: empty vector infected control groups.
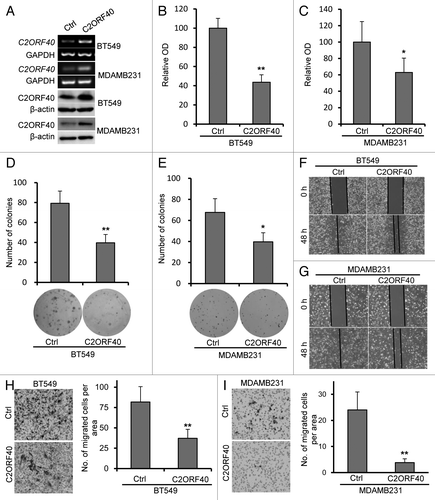
Figure 4. Knockdown of C2ORF40 promotes breast cancer cell growth and migration-invasion. (A) Knockdown of C2ORF40 in SKBR3 and MDAMB468 cells as indicated by RT-PCR (upper panel) and western blotting (lower panel). Knockdown of C2ORF40 promotes SKBR3 and MDAMB468 cell proliferation assessed by MTT method (B, C). Representative photographs show knockdown of C2ORF40 promotes migration of SKBR3 (D) and MDAMB468 (E) cells in a scratch-wound healing model on cultured cells. Knockdown of C2ORF40 promotes the invasive ability of SKBR3 (F) and MDAMB468 (G) cells using Matrigel assay. Images in (D) to (G) show representative field of views. All experiments were repeated at least three times. Data were presented as mean ± SD, * p < 0.05 and ** p < 0.01 obtained from Student’s t-test as compared with control groups. shCtrl: scrambled vector infected control groups.
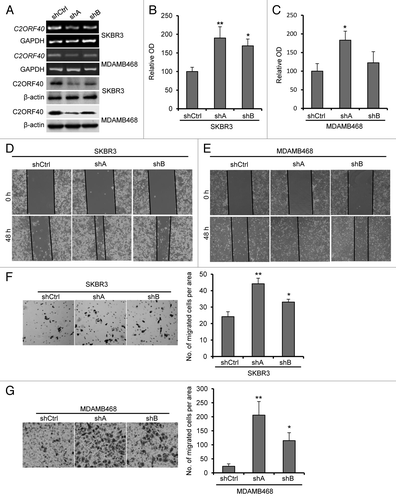
Figure 5.C2ORF40 regulates mitosis-associated gene expression and M phase progression of cell cycle. IPA networks of genes significantly correlated with C2ORF40 invoked cell cycle, cellular assembly and organization, DNA replication, recombination and repair (A) and cellular movement, cancer, cardiovascular system development and function (B). GO terms associated with lists of genes that are significantly correlated with C2ORF40(C). Cell cycle progression in control vector (left panels) and C2ORF40 (right panels) transfected BT549 (D) and MDAMB231 (E) cells was determined by FACS Caliber cytometry. There is a significant increase in G2/M cell population in the cells with enforced expression of C2ORF40(F). The experiments in (D to F) were repeated at least three times in two independently retrovirally transduced cell lines and data in (F) were presented as mean ± SD P-values shown in (F) were obtained from Student’s t-test. * p < 0.05 and ** < 0.01. Ctrl: empty vector infected control groups.
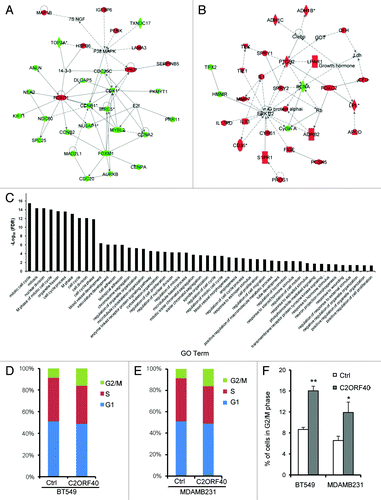
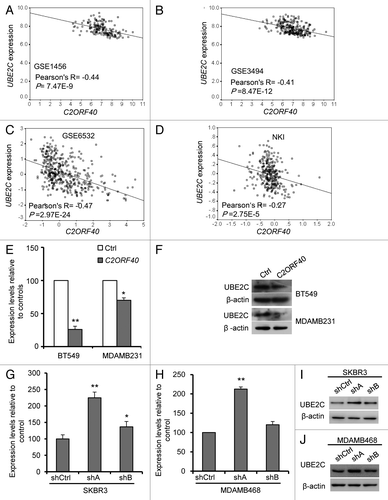
Figure 6.C2ORF40 suppresses UBE2C expression. There is a highly significant and negative correlation between C2ORF40 and UBE2C in mRNA levels within human breast cancer tissues from GEO database Data set 5 (A), Data set 6 (B), Data set 7 (C), Data set 4 (D). Ectopic expression of C2ORF40 in BT549 and MDAMB231 cells downregulates the mRNA (E) and protein (F) levels of UBE2C. Knockdown of C2ORF40 in SKBR3 (G and I) and MDAMB468 (H and J) cells increases the mRNA (G and H) and protein (I and J) levels of UBE2C. The experiments in (E) to (J) were repeated at least three times and data were presented as mean ± SD. R is Pearson correlation coefficient. P-values shown in (A to D) were obtained from Pearson correlation test and in (E), (G) and (H) were obtained from Student’s t-test. * p < 0.05 and ** < 0.01. Ctrl: empty vector infected control groups, shCtrl: scrambled vector infected control groups.