Figures & data
Figure 1. LOS bovine fetuses have similar phenotype characteristics as those reported in BWS patients. (A). Fetal weight at day ~105 gestation. Y axis represents the weight in grams. X axis has no actual implication and is used to scatter the spots representing each fetus for ease of visualization. The sex of the fetuses and the way they were generated is shown at the top-right side. The bold line represents the 97th percentile of control weight (i.e. 476.8 g). (B). Primary and secondary characteristics of BWS can be observed in LOS; B.1 = AI-B884 (control female weighing 400 g) and B.2 = AI-B799 (control male weighing 408 g which is the approximate average weight of the control fetuses). B.3 and B.4 show fetuses with macrosomia (ART-J835LOS – female weighing 714 g and ART-J489ALOS– female weighing 514 g). B.5 shows an example of macroglossia in a female weighing 620 g and B.6 shows an ear malformation in a female weighing 320 g. Each square on the background = 2.54 cm2. LOS, large offspring syndrome; BWS, Beckwith-Wiedemann syndrome; AI, artificial insemination; ART, assisted reproductive technologies.
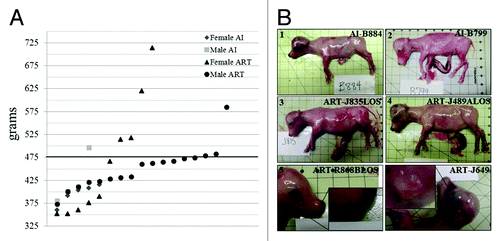
Figure 2. Example of assays used to determine allelic expression in tissues from B. t. indicus × B. t. taurus F1 hybrid conceptuses. Shown are examples of allelic determination by RT-PCR followed by RFLP and PAGE (A-C), Sanger sequencing (D) or SSCP analysis (E-F). The left portion of the panels (A-C, E and F) shows the band pattern of B. t. taurus and B. t. indicus control tissues (liver) which was used as reference to determine parental expression of imprinted gene in tissues from B. t. indicus × B. t. taurus F1 hybrids. The right portion of the panels shows examples of monoallelic and biallelic expression of several imprinted genes in ~d105 conceptus. (D) is an example of the Sanger sequencing allelic assay for KCNQ1OT1, a paternally-expressed gene. Two SNPs were used in this assay; double peaks demonstrate biallelic expression. The contribution of each parental allele to the total expression was determined by the use of Image J (NIH). Only samples with at least 10% expression from the repressed allele were considered to be biallelically-expressed. T, B. t. taurus; i, B. t. indicus; mono, monoallelic; bi, biallelic; SNP, single nucleotide polymorphism; RFLP, restriction fragment length polymorphism; SSCP, single strand conformation polymorphism; PAGE, polyacrylamide gel electrophoresis.
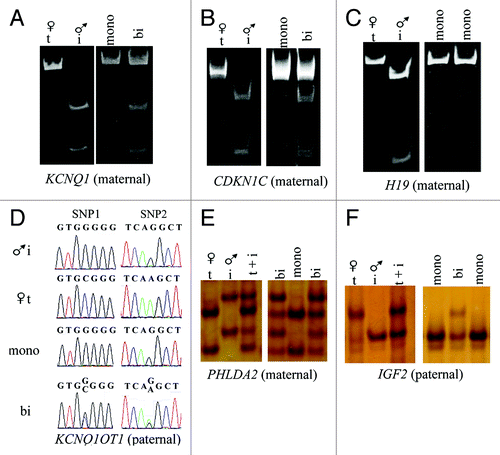
Figure 3. Biallelic expression of KCNQ1OT1 in LOS fetuses. Shown is Sanger sequencing data of KCNQ1OT1 RT-PCR product in tissues analyzed in AI-B799 (control), ART-J835LOS and ART-J489ALOS fetuses. The columns show the chromatograph for each tissue of each fetus. Values below the chromatograph are the percentage of KCNQ1OT1 expressed from the maternal allele. Arrows show the double peaks of SNP1 site (refer to ) in ART-J835LOS and ART-J489ALOS fetuses. For clarity of depiction, SNP2 site is shown here (Fig. S1). LOS, large offspring syndrome; RT-PCR, reverse transcription-polymerase chain reaction; SNP, single nucleotide polymorphism.
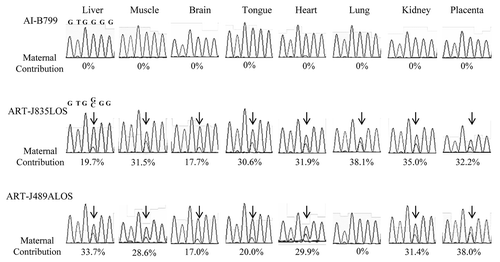
Figure 4. LOS fetuses with biallelic expression of KCNQ1OT1 show downregulation of CDKN1C. (A) The CDKN1C RNA was determined by quantitative RT- PCR in several tissues from eight AI fetuses (diamonds; AI), two LOS fetuses with biallelic expression of KCNQ1OT1 (triangle = ART-J835LOS; square = ART-J489ALOS; bi-LOS), and five LOS fetuses with correct imprinting of KCNQ1OT1 (circles; mono-LOS). The short line among the diamonds and circles represents the average level of the individuals. CDKN1C level was normalized to the expression of GAPDH. Note that for each tissue analyzed CDKN1C level in ART-J835LOS is at least 2-fold lower than the average level found in AI fetuses and LOS fetuses with correct imprinting of KCNQ1OT1. It should be also noted that lung from ART-J489ALOS which had correct imprinting of KCNQ1OT1 () exhibits the comparative CDKN1C level with controls. (B) The threshold cycle (Ct) of CDKN1C was normalized to the reference gene GAPDH in each tissue from the eight AI fetuses. The data are expressed as mean ± SEM. *p < 0.05 between the brain, heart and other tissues analyzed. LOS, large offspring syndrome.
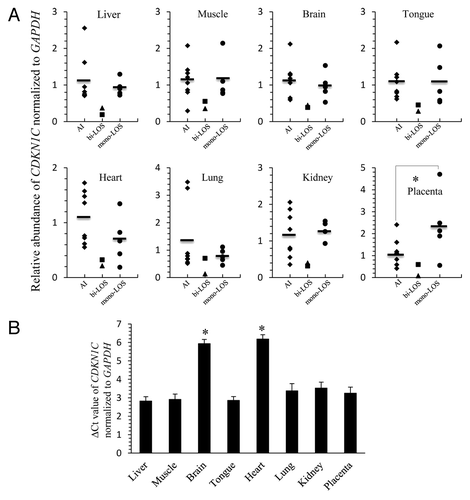
Figure 5. Loss of methylation of KvDMR1 on the maternal allele is associated with biallelic expression of KCNQ1OT1 in LOS fetuses. DNA was treated with sodium bisulfite prior to PCR, and PCR product was cloned before sequencing. Sequencing data was used to determine the DNA methylation status at the KvDMR1. Shown on top is a depiction of the 10th intron of the maternally-expressed gene KCNQ1 and its direction of transcription is shown with an arrow. The region harbors the promoter of the antisense long ncRNA KCNQ1OT1 (shown as dashed arrow), which is also an imprinting control region known as KvDMR1. A 385 bp region of the KvDMR1 was used to determine the DNA methylation status of 37 CpG sites (ovals). A SNP (vertical arrow) between B. t. indicus and B. t. taurus was used to determine the parental origin of the alleles and only maternal alleles are shown here. Five tissues from two fetuses are shown. Filled and open circles represent methylated and unmethylated CpG dinucleotides, respectively. Missing circles are sequencing data of low quality. Each line denotes an individual DNA strand. The level of maternal KCNQ1OT1 expression is shown in the center and next to the strands. Tail tissues were collected for the purpose of DNA analysis, precluding its use for gene expression determinations. NA, not available.
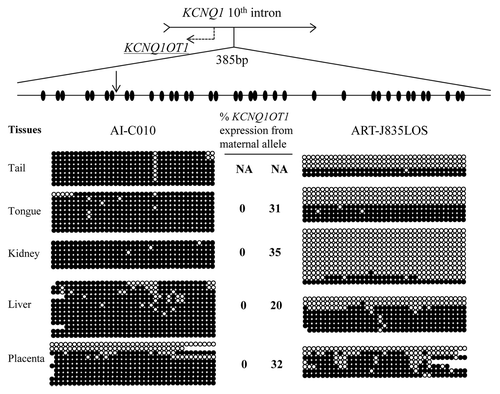
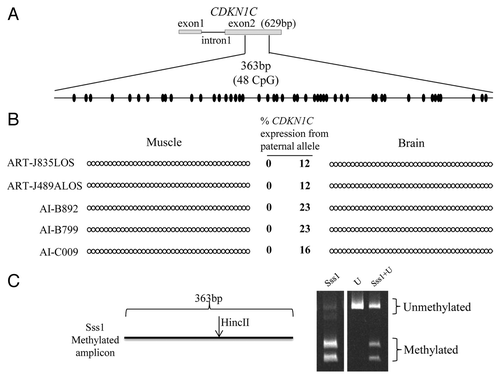
Figure 6. Imprinted expression of CDKN1C is not regulated by somatic DNA methylation. (A) Shown on top is a depiction of the first two exons and the first intron of the maternally-expressed gene CDKN1C. No DNA polymorphisms were identified in this region. Bisulfite converted specific primers for a 363 bp region of the exon 2 were used to determine the DNA methylation status of 48CpG sites (ovals). (B) The PCR product was sequenced without cloning. Two tissues from five fetuses are shown. Filled and open circles represent methylated and unmethylated CpG dinucleotides, respectively. Each line denotes the DNA methylation pattern of each sample. Only the first 39 CpGs are shown here because of the low quality sequencing of last 9 CpGs due to primer binding. The level of paternal CDKN1C expression is shown in the center and next to the strands (based on a SNP in exon 4). (C) COBRA performed to ensure ability of primers to equally amplify methylated and unmethylated alleles. The restriction enzyme HincII recognized and cleaved the methylated amplicon. Sss1 = genomic DNA co-incubated with the methyltransferase Sss1 prior to bisulfite conversion, U = bisulfite converted DNA with no Sss1 treatment, Sss1 + U = equal portions of methylated and unmethylated DNA were used for PCR amplification. The digested products were resolved by PAGE.